With recent advances in genome sequencing technology, a large body of evidence has accumulated over the last few years linking alterations in microbiota with cardiovascular disease. In this study, we aimed to compare gut microbial composition using 16S ribosomal DNA (rDNA) sequencing techniques in patients with coronary artery disease (CAD) and stable heart failure (HF) with reduced ejection fraction and patients with CAD but with normal ejection fraction. We also studied the relationship between systemic inflammatory markers and microbial richness and diversity.
MethodsA total of 40 patients (19 with HF and CAD, 21 with CAD but without HF) were included in the study. HF was defined as left ventricular ejection fraction <40%. Only stable ambulatory patients were included in the study. Gut microbiota were assessed from the participants’ fecal samples. The diversity and richness of microbial populations in each sample were assessed by the Chao1-estimated OTU number and the Shannon index.
ResultsThe Chao1-estimated OTU number and Shannon index were similar between HF and control groups. There was no statistically significant relationship between inflammatory marker levels (tumor necrosis factor-alpha, interleukin 1-beta, endotoxin, C-reactive protein, galectin-3, interleukin 6, and lipopolysaccharide-binding protein) and microbial richness and diversity when analyzed at the phylum level.
ConclusionIn the current study, compared to patients with CAD but without HF, stable HF patients with CAD did not show changes in gut microbial richness and diversity. At the genus level Enterococcus sp. was more commonly identified in HF patients, in addition to certain changes in species levels, including increased Lactobacillus letivazi.
Os recentes avanços na tecnologia de sequenciação de genoma permitiram gerar conhecimento que possibilite relacionar alterações da microbiota com doenças cardiovasculares. Neste estudo, comparámos a composição microbiana intestinal, utilizando técnicas de sequenciação do DNA ribossómico 16S (rDNA), em doentes com fração de ejeção reduzida estável com insuficiência cardíaca (IC) e com doença arterial coronária (DAC) e em doentes com DAC, mas com fração de ejeção normal. Estudámos ainda a relação com marcadores inflamatórios sistémicos e a riqueza e diversidade microbiana.
MétodosForam incluídos no estudo 40 pacientes (19 com insuficiência cardíaca e DAC, 21 com DAC). A insuficiência cardíaca foi definida como fração de ejeção do ventrículo esquerdo <40%. Apenas doentes de ambulatório estáveis foram incluídos no estudo. A microbiota intestinal foi avaliada a partir de amostras fecais dos participantes. A diversidade e riqueza das populações microbianas em cada amostra foi avaliada pelo número de OTU estimado de Chao1 e índice de Shannon.
ResultadosO número de OTU estimado de Chao1 e o índice de Shannon foram semelhantes entre os grupos IC e controlo. Não houve relação estatisticamente significativa entre os níveis de marcadores inflamatórios (fator de necrose tumoral-alfa, interleucina 1-beta, endotoxina, proteína C-reativa, galectina-3, interleucina-6 e proteína de ligação a lipopolissacarídeos) e a riqueza e diversidade microbiana quando analisados ao nível do filo.
ConclusõesNo presente estudo, comparados aos doentes com DAC, mas sem DAC, os doentes com IC estável com DAC não apresentaram as alterações na riqueza e diversidade microbiana intestinal. No nível de gênero Enterococcus sp. foi maior nos doentes com IC, além de algumas alterações nos níveis de espécies, incluindo aumento de Lactobacillus letivazi.
The human body is inhabited by a diverse and complex microbial community that lives in coexistence with its host and is referred to as the microbiota. The microbiota mainly colonize the gastrointestinal tract, the colon harboring the densest number of microbes.1 Although it is known that the intestinal microbiome plays an essential role in the maintenance of host homeostasis and physiology, until recently information on human microbiota and their interactions with the host was limited, mainly due to technological shortcomings. However, recent advances in genome sequencing technologies and bioinformatics have enabled researchers to study microorganisms and their potential role in cardiovascular disease in detail.2 Subsequently, a large body of evidence has accumulated over the last few years linking alterations of intestinal microbiota with cardiovascular and metabolic disease, including coronary artery disease (CAD) and heart failure (HF).3
HF is a multisystem disorder that affects not only the heart and circulation but also intestinal function. Depressed cardiac function leads to decreased intestinal perfusion, mucosal edema, and local pH changes in the intestinal lumen.4,5 Together with a lack of immunological defense, an increased mucosal bacterial biofilm area can be seen in HF patients compared to non-HF patients.4 Moreover, the villous structure of the intestinal wall is also affected, leading to impaired integrity of the intestinal barrier, which consequently promotes the translocation of bacteria, bacterial fragments such as endotoxin, and secreted products into the circulatory system.6 Previous studies showed that higher endotoxin levels induce immune system activation and cytokine production, contributing to the chronic inflammation present in HF.7,8 Altered microbial composition has also been shown in some other studies.9–11
All of these results illustrate the complex interplay between microbiota and host in HF patients. However, it is unclear whether the presence of HF per se, rather than comorbidities including CAD and diabetes, is responsible for microbial alterations. In this study, we aimed to use 16S ribosomal DNA (rDNA) sequencing techniques to compare the gut microbial composition in stable systolic HF patients with CAD and patients with CAD but without HF. To study a more homogenous group and to minimize the effect of potential confounders, we studied only stable patients with CAD. Moreover, we explored the relationship between the microbiota and systemic inflammatory markers, which included tumor necrosis factor-alpha (TNF-α), interleukin 1-beta (IL-1β), endotoxin, C-reactive protein (CRP), galectin-3 (Gal-3), interleukin 6 (IL-6), and lipopolysaccharide-binding protein (LBP).
MethodsThe study was carried out in patients aged between 40 and 74 years who were treated at cardiology outpatient clinics in Pamukkale University Hospital, Turkey. Nineteen patients with a diagnosis of HF and CAD were allocated to the patient group, while 21 patients with CAD but without HF served as the control group. HF was defined as a left ventricular ejection fraction of <40% measured by the modified Simpson method on echocardiography and at least one symptomatic HF presentation. Patients with symptoms of active infection, a chronic inflammatory disease diagnosis, malignancy, diabetes, renal failure or renal replacement, a history of gastrointestinal surgery, and patients receiving immunosuppressive agents including antibiotics, probiotics, or steroids in the previous two months, were excluded from the study. Written informed consent was obtained from each participant. The study was approved by the institutional ethics committee.
Enzyme-linked immunosorbent assaysHuman TNF-α, human IL-1β, human endotoxin, human CRP, human Gal-3, human IL-6, human LBP, and human N-terminal brain natriuretic peptide (NT-proBNP) enzyme-linked immunosorbent assay (ELISA) kits were applied to serum samples taken from the patient and control groups, and analyses of the samples and determination of concentrations were performed by the sandwich-based ELISA method. Except endotoxin tests (REL, Turkey), all kits were purchased from Elabscience, China. All tests were performed according to the manufacturer's instructions. The absorbances of serum samples in the 96-well plate were measured at 450 nm using a Heales MB-530 microplate reader. The concentrations of protein in each well were calculated using the kit's standard equations. All results of concentration levels were assessed in the control and study groups.
16S ribosomal DNA sequencingFecal samples of participants were freshly collected, transported to the laboratory under anaerobic conditions at 4°C, frozen with liquid nitrogen in phosphate buffered saline containing 20% glycerol, and stored at −80°C until use. Bacterial genomic DNA was extracted from the fecal samples using a genomic DNA extraction kit (PureLink® Genomic DNA Mini Kit, Thermo Fisher, USA).
The partial 16S ribosomal RNA (rRNA) from the bacterial genomic DNA was amplified by polymerase chain reaction (PCR), as previously described.17 Primers 27F-mod and 338R with adapter sequences were used to amplify the bacterial V1-V2 region of the 16S rDNA gene. Each 25-μl PCR reaction mix contained 25 ng/μl microbial genomic DNA, 1 μM of each primer and 12.5 μl 2×KAPA HiFi HotStart ReadyMix (Roche, Switzerland). The PCR conditions for DNA were as follows: initial denaturation for 5 min at 95°C; 30 cycles of 15 s at 95°C, 15 s at 42°C and 30 s at 72°C; and 72°C for 5 min for final extension. Agencourt AMPure XP magnetic beads (Beckman Coulter, UK) were used to purify the amplicons. A subsequent limited-cycle amplification step was performed to add multiplexing indices and Illumina adapter sequences i5 and i7. Amplicons were quantified, normalized and pooled using the Qubit® 1X dsDNA HS assay kit (Life Technologies, Thermo Fisher, US). Library preparation was carried out to prepare 2×300 base-pair sequencing on the Illumina MiSeq platform to obtain the longest possible reads.
The sequences were analyzed using the previously established pipeline.17 Filtered reads with a mean quality score of 25 or higher were randomly selected from the reads for each sample. The high-quality reads of each sample were clustered at a 96% identity threshold in operational taxonomic units (OTUs). The read sequences were aligned against the 16S rRNA gene database constructed from the Ribosomal Database Project, CORE, and NCBI genome databases, and were assigned to taxonomic groups at a 96% identity threshold.
Statistical analysisStatistical analysis was performed using PSPP 1.0.1 and R (with the EasyR plugin). A p-value less than 0.05 was considered significant. Continuous variables were summarized as mean±standard deviation and categorical variables as count and percentages in the tables. Continuous variables were investigated using analytical methods (Kolmogorov-Smirnov and Shapiro-Wilk tests) to determine normality of distribution. One-way analysis of variation was used to compare continuous variables with normal distribution and the Kruskal-Wallis test was used to compare variables with non-normal distribution. Relationships between categorical variables were analyzed by the chi-square test.
Real-time polymerase chain reaction analysis of the findings was performed using the ΔΔCT method and quantified with a computer program. The groups were compared using volcano plots from RT2-Profiler™ PCR Array Data Analysis, assessed statistically using the Student's t test.
Microbiota analyses were performed with R software (version 3.5.1, GPL-3 license). Microbial diversity was assessed at kingdom, phylum, class, order, family, genus and species level. The richness and diversity of microbial populations in each sample were assessed via Chao1-estimated operational taxonomic unit (OTU) number and the Shannon index.
ResultsThe demographic characteristics of the study population are shown in Table 1. The groups were comparable in terms of age, body mass index (BMI), smoking, and hypertension status. Males were predominant in similar proportions in both groups. IL-1β, TNF-α, and CRP levels were significantly different between the two groups (Table 2).
Baseline characteristics of the study population.
| Heart failure (n=19) | Controls (n=21) | p | |
|---|---|---|---|
| Male | 17 (89.4%) | 19 (90.4%) | 1.000 |
| Age, years | 56.38±7.86 | 54.76±7.60 | 0.516 |
| BMI, kg/m2 | 29.03±2.62 | 30.11±2.46 | 0.196 |
| Fasting glucose, mg/dl | 94±10.50 | 98±19.00 | 0.053 |
| TC, mg/dl | 156±71.75 | 157±42 | 0.955 |
| HDL-C, mg/dl | 38.33±9.92 | 37.00±7.30 | 0.633 |
| LDL-C, mg/dl | 99.38±37.51 | 91.47±31.53 | 0.479 |
| Triglycerides, mg/dl | 160.16±56.06 | 160.38±37.74 | 0.989 |
| Creatinine, mg/dl | 0.99±0.17 | 0.88±0.26 | 0.114 |
| Smoking | 8 (44%) | 6 (28%) | 0.487 |
| Hypertension | 4 (22%) | 4 (19%) | 1.000 |
| LVEF, % | 31.4±8.25 | 55.2±10.00 | <0.001 |
| Medications | |||
| Aspirin | 17 (94%) | 20 (95.2%) | 1.000 |
| P2Y12 inhibitor | 7 (38%) | 17 (0.9%) | 0.018 |
| Beta-blocker | 15 (83%) | 20 (95.2%) | 0.318 |
| Statin | 11 (61%) | 19 (90.4%) | 0.055 |
| ACEI/ARB | 15 (83%) | 11 (52.3%) | 0.088 |
| PPI | 13 (72%) | 13 (61.9%) | 0.733 |
| Spironolactone | 10 (55%) | 0 (0%) | <0.001 |
| Furosemide | 13 (68%) | 7 (33%) | 0.028 |
ACEI/ARB: angiotensin-converting enzyme inhibitor and/or angiotensin receptor blocker; BMI: body mass index; HDL-C: high-density lipoprotein cholesterol; LDL-C: low-density lipoprotein cholesterol; LVEF: left ventricular ejection fraction; PPI: proton pump inhibitor; TC: total cholesterol.
Continuous variables were summarized as mean±standard deviation and categorical variables as count and percentage.
Comparison of blood inflammatory markers between the groups.
| Total (n=40) | Heart failure (n=19) | Controls (n=21) | p | |
|---|---|---|---|---|
| Gal-3, ng/ml | 7.39±0.34 | 7.34±0.31 | 7.44±0.34 | 0.89 |
| IL-1β, pg/ml | 1.49±0.16 | 1.52±0.21 | 1.47±0.03 | <0.05 |
| IL-6, pg/ml | 2.37±0.86 | 2.37±0.44 | 2.50±1.08 | 0.76 |
| CRP, ng/ml | 6.22±1.63 | 6.78±1.5 | 5.60±1.53 | <0.05 |
| Endotoxin, EU/l | 6.56±1.79 | 6.54±1.5 | 6.58±2.05 | 0.56 |
| LBP, ng/ml | 12.01±3.03 | 12.23±3.07 | 11.81±3 | 0.91 |
| TNF-α, pg/ml | 1.99±0.9 | 2.39±0.25 | 1.63±1.14 | <0.01 |
| NT-proBNP, ng/ml | 11.54±4.47 | 12.21±3.31 | 10.94±5.23 | 0.82 |
CRP: C-reactive protein; Gal-3: galectin-3; IL-1β: interleukin-1 beta; IL-6: interleukin 6; LBP: lipopolysaccharide binding protein; NT-proBNP: N-terminal pro-brain natriuretic peptide; TNF-α: tumor necrosis factor alpha.
Continuous variables were summarized as mean±SD.
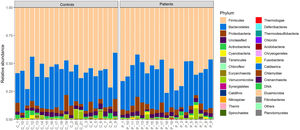
16S rRNA gene sequencing was carried out on the fecal samples of the 19 patients with HF and 21 controls. The majority of the intestinal microbiota were dominated by three phyla, Firmicutes, Bacteroidetes, and Proteobacteria. At all levels of taxa, the gut microbiota were similar between patients and controls (Figure 1).
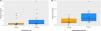
The diversity and richness of microbial populations in each sample were assessed by the Chao1-estimated OTU number and the Shannon index. No differences were found at phylum, genus, or species levels between the groups (Figure 2).
A Kruskal-Wallis test revealed that there were no statistically significant differences between HF patients and the control groups for Gal-3 (p=0.46), IL-1β (p=0.67), IL-6 (p=0.13), CRP (p=0.24), endotoxin (p=0.33), LBP (p=0.11) or TNF-α (p=0.48).
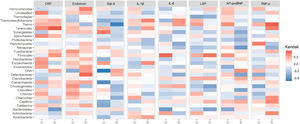
As shown in Figure 3, the Kendall rank correlation coefficient showed no statistically significant relationship between inflammatory marker levels and microbial richness and diversity when analyzed at the phylum level.
Distribution of rank correlation coefficients between patients (P) and controls (C) at phylum level. CRP: C-reactive protein; IL-1β: interleukin 1-beta; IL-6: interleukin 6; Kendall: Kendall rank correlation coefficient; LBP: lipopolysaccharide binding protein; NT-proBNP: N-terminal pro-brain natriuretic peptide; TNF-α: tumor necrosis factor alpha.
In the assessment of beta diversity, no significant difference was observed between patients and controls according to Bray-Curtis dissimilarity based on genera abundance.
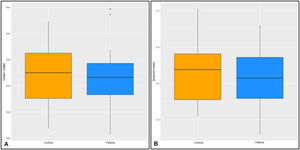
The Promicromonosporaceae family (Figure 4A) in the phylum Actinobacteria and the Flammeovirgaceae family (Figure 4B) in the phylum Bacteroidetes were less often identified in HF patients than in the control group (p<0.05).
At the genus level, Enterococcus spp. was found at higher levels in the patient group than in controls (p=0.03) (Figure 5A). At the species level, Lactobacillus letivazi was found to be significantly increased in samples taken from patients compared to the control group (p=0.008) (Figure 5B). In addition, Providencia burhodogranariea, Bifidobacterium angulatum, B. gallicum, Peptoniphilus gorbachii, and Anoxybacillus ayderensis were found to be significantly decreased (p<0.01) in HF patients at species levels compared to the control group.
DiscussionThe results of the current study are twofold. First, using 16S rRNA gene sequencing, we found no statistically significant difference between gut microbiota richness assessed in fecal samples from stable, compensated HF patients with CAD and those from patients with CAD but without HF. Secondly, inflammatory markers including TNF-α, IL-1β, endotoxin, CRP, Gal-3, IL-6, and LBP were not correlated with indices of microbial richness in stable CAD patients with or without HF.
The gut microbiome varies significantly among healthy individuals, and there is no general consensus on which features constitute a healthy microbiome.12–14 Two main phyla of bacteria, Firmicutes and Bacteroidetes, are predominant, accounting for more than 90% of total bacteria, and their ratio is usually considered a relative signature for health status, although this is still controversial.13 Alternatively, the richness and diversity of the microbiota can be assessed with indices such as the Chao1 and Shannon indices.
Only a limited number of studies have examined the gut microbiota in HF. Pasini et al. studied the intestinal overgrowth of Candida and pathogenic bacteria in HF patients and found increased development rates of the pathogens in patients with moderate to severe New York Heart Association (NYHA) functional classes compared to patients with mild NYHA functional classes.15 Using 16S rRNA gene sequencing, Luedde and colleagues investigated the intestinal microbiome of 20 patients and showed that compared to controls, HF cases had a significantly decreased alpha (intra-individual) diversity.9 Moreover, beta (inter-individual) diversity measures yielded a highly significant separation between HF cases and controls. The authors also reported significant decreases in Coriobacteriaceae, Erysipelotrichaceae, and Ruminococcaceae at the family level and significant decreases in Blautia, Collinsella, unclassified Erysipelotrichaceae, and unclassified Ruminococcaceae in HF cases compared to controls at the genus level.9 In a subsequent study, Cui et al. performed metagenomic and metabolomic analyses of fecal samples from patients with chronic HF compared to controls and found that the composition of gut microbiota in HF was significantly different from that of controls.10 Additionally, decreased Faecalibacterium prausnitzii and increased Ruminococcus gnavus were the essential characteristics in the gut microbiota of patients with HF.10 Kamo et al. analyzed the differences in gut microbiota in fecal samples from 12 HF patients and 12 age-matched healthy controls. Although the richness and diversity of gut microbiota were similar between the samples from HF patients and control subjects according to the Chao1 and Shannon indices, the composition of the gut microbial communities of HF patients was different from that of controls in both unweighted and weighted UniFrac analyses.16 Additionally, Eubacterium rectale and Dorea longicatena were less abundant in the gut microbiota of HF patients.16 In another Japanese study, researchers assessed stool samples from 28 non-ischemic HF patients with low ejection fraction and 19 healthy controls.17 While alpha diversity did not differ between groups, ordination by beta-diversity metrics revealed a separation of the groups across components of variation. Streptococcus and Veillonella were enriched in the common core microbiota of patients, while SMB53 was depleted.17 Kummen and colleagues studied two independent cohorts of stable systolic HF patients (discovery, n=40 and validation, n=44), all of whom were in NYHA class II–IV for more than six months, and population-based control subjects. They found that in both cohorts, patients with HF had decreased microbial richness, even after adjustment for age, gender, BMI, hypertension, and diabetes. Investigators also identified alterations in 15 taxa in both cohorts, and the majority of depleted taxa, including taxa in the Lachnospiraceae family, had butyrate-producing potential.11
There are several discrepancies, however, in terms of the major bacterial groups identified in the studies.18 Similarly, diversity indices are inconsistent between studies. However, our results were in agreement with the study by Kamo et al., in which they did not show differences in alpha- or beta- diversity metrics.16 A recent study in rats also showed similar results to our study. Lataro and colleagues studied the gut microbiota of rats in fecal samples collected before and six weeks after left anterior descending artery ligation or sham surgery.19 The investigators failed to show changes in species richness and diversity of the gut microbiota, the relative abundance of phyla, the number of species in each phylum and the microbial community, or the acetate-, butyrate- or lactate- producing bacterial populations in rats with HF compared to the sham arm.
Moreover, there are other possible explanations for the discrepancy between our study and others. First, in this study, which aimed to reduce the effect of potential confounders, we only included HF patients with CAD in the patient group, and compared them with patients with CAD but without HF as controls. This was one of the main differences between our analysis and previous studies. Furthermore, we also excluded patients with diabetes and renal failure, because the dietary habits or medications of these patients can affect the composition of gut microbiota, or the disease, per se, can be a confounder.20–23 Second, we only included stable, compensated HF patients in the study. Some studies suggest that alterations in the gut and in gut microbiota may only be noticeable in acute decompensated states or severe HF. For instance, the composition of intestinal microbiota was shown to change dynamically during intestinal ischemia and reperfusion in rats.24 Acute venous congestion of the gut can lead to increased gut permeability for bacteria, endotoxin, or both, and the increased translocation of these materials into the bloodstream.8 Niebauer et al. showed significantly increased concentrations of endotoxin and cytokines in patients with chronic HF during an acute edematous exacerbation, which normalized with intensified diuretic treatment.8 In the above-mentioned study by Pasini et al., right atrial pressure, an indicator of venous congestion, was correlated with intestinal permeability (r=0.55, p<0.0001).15 In the study by Luedde et al., 70% of the patients were in an acute state of cardiac decompensation, and only 30% were in a stable state of HF.9 Similarly, patients included in the study by Cui et al. were in-hospital patients, who were admitted because of their unstable illness status, with the majority having poor functional capacity (51% in NYHA class III, 43% in class IV, 6% in class II, and none in class I).10 Consequently, patients’ volume status or state of decompensation could be a driver for the discrepancy between the studies. Finally, various demographic and environmental factors, including diet and medications, can influence the gut microbial composition and lead to inconsistent results.25,26 For example, a recent study of the above-mentioned HF cohorts related gut microbial alterations at least partly to low fiber intake, emphasizing the importance of diet.27
In the present study, there was no relationship between microbial diversity in the gut and inflammatory markers. However, in addition to inflammation, microbiota can interact with the host through many other pathways, including metabolic pathways. In fact, microorganisms in the gut can generate various microbial metabolites, including trimethylamine N-oxide (TMAO), short-chain fatty acids, tryptophan metabolites, and bile acid metabolites.28 In particular, the metaorganismal molecule TMAO, which is the hepatic oxidation product of the microbial metabolite trimethylamine, has recently attracted considerable attention as a potential mediator of cardiovascular disease.28,29 Previous studies demonstrated that TMAO was not only a prognostic marker in HF, but can also directly affect cardiac performance by inducing myocardial hypertrophy, fibrosis, inflammation and mitochondrial dysfunction.30–32
LimitationsWe did not include any measurements of microbial metabolites in our study, which can be considered as a limitation. The complex interaction between gut microbiota and HF/CAD warrants further research. A second limitation of our study is the small sample size. Thirdly, subjects included in this study were all from the same city, aged between 40 and 74 years, and were consuming foods typically found in the local diet. The lack of objective questionnaires to document their dietary habits was another limitation of this study. Fourth, sequential patient enrollment resulted in male predominance in the groups, which can at least be partially explained by the higher frequency of CAD in males, even in the absence of other risk factors. Finally, including another patient group with acute decompensated HF with measurements of gut epithelial barrier function and blood flow would have helped to explain some of the discrepancies between the studies and would have provided more insights into the relevant pathophysiology.
ConclusionIn patients with HF, the composition of the gut microbiota can be influenced by both host genotype and lifestyle factors such as diet, physical activity, medications, comorbidities, and volume status. Compared to CAD patients without HF, stable HF patients with CAD did not show changes in microbial richness and diversity, indicating stable HF per se may not affect the gut microbial composition. However, at the genus level Enterococcus sp. was more commonly identified in HF patients, in addition to certain changes in the species levels, including increased Lactobacillus letivazi. The effect of HF on gut microbiota requires further research.
Conflicts of interestThe authors have no conflicts of interest to declare.
This study was supported by the Pamukkale University Scientific Research Projects Coordination Unit, dated 17.01.2018, decision number 2018TIPF005.