Genetic susceptibility has a key role in the pathogenesis of coronary artery disease (CAD). KLF5 and KLF7 are transcriptional factors essential to cell development and differentiation. Their genetic variants have been associated with the risk of metabolic disorders. The present study aimed to evaluate the possible correlation of KLF5 (rs3812852) and KLF7 (rs2302870) single nucleotide polymorphisms (SNPs) with the risk of CAD for the first time in the world.
MethodsThe clinical trial study comprised 150 patients with CAD and 150 control subjects without CAD from the Iranian population. After blood sampling, deoxyribonucleic acid was extracted and genotyped using the Tetra Primer ARMS-PCR method and confirmed by Sanger sequencing.
ResultsThe KLF7 A/C genotypes and C allele frequency were meaningfully higher in the control group compared to the CAD+ group (p<0.05). No obvious association has been observed between KLF5 variants and CAD risk. However, the distribution of the AG genotype of KLF5 was statistically lower in CAD+ patients with diabetes than in CAD+ patients without diabetes (p<0.05).
ConclusionThis study identified KLF7 SNP as a causative gene contributing to CAD, which presents novel insight into the molecular pathogenesis of the disease. It is, however, unlikely that KLF5 SNP has an essential role in the risk of CAD in the studied population.
A suscetibilidade genética tem um papel fundamental na patogénese da doença arterial coronária (DC). O KLF5 e o KLF7 são fatores de transcrição que desempenham papéis importantes no desenvolvimento e diferenciação celular e as suas variantes genéticas têm sido associadas ao risco de distúrbios metabólicos. O presente estudo teve como objetivo avaliar a possível correlação dos polimorfismos do nucleotídeo único (SNPs) KLF5 (rs3812852) e KLF7 (rs2302870) com o risco de DC pela primeira vez no mundo.
MétodosO estudo de ensaio clínico compreende 150 pacientes com DC (CAD+) e 150 casos controlos sem DC na população iraniana. Após colheita de sangue, o DNA foi extraído e genotipado pelo método Tetra Primer ARMS-PCR, confirmado pela sequência de Sanger.
ResultadosOs genótipos KLF7 A/C e a frequência do alelo C foram significativamente maiores no grupo controlo em comparação com o grupo CAD+ (p<0,05). Nenhuma associação óbvia foi observada entre as variantes do KLF5 e o risco de CAD+. No entanto, a distribuição do genótipo AG de KLF5 foi estatisticamente menor em doentes CAD+ com diabetes do que em doentes CAD+ sem diabetes (p<0,05).
ConclusãoEste estudo identificou o SNP KLF7 como um gene causador da DC, que apresenta uma nova visão sobre a patogénese molecular da doença. Mas é improvável que o SNP KLF5 tenha um papel essencial no risco de DC na população estudada.
Coronary artery disease (CAD) is the most common cardiovascular abnormality and the main cause of human death around the globe. CAD is an atherosclerotic injury with an inflammatory basis, induced when an obstruction (more than 50%) is created in at least one of the coronary arteries.1 Generally, CAD is common among the elderly. However, recent studies have demonstrated that 4–10% of patients who suffer from CAD are <45 years old. The World Health Organization report, presented in 2009, revealed that 17.3 million deaths are caused by cardiovascular diseases (CVD). In 2016, the American Heart Association reported that 15.5 million people >20 years old suffer from CAD complications, and approximately 500000 deaths accrue annually due to CAD.2 CAD-related mortality in Iran is approximately 175000 annually, representing 50% of all deaths.3
Besides environmental factors and lifestyle, family history and several genetic polymorphisms are strongly associated with CAD. Therefore, the mutation in the genes linked to these diseases could be useful in distinguishing the disease vulnerability of a general population. Genome-wide association analysis of CAD determined that 396 single-nucleotide polymorphisms (SNPs) correlated with CAD. Of these, 30 SNPs were located in nine chromosomes.4
KLFs are zinc-finger-containing transcriptional controllers that mediate important cellular processes such as development, growth, proliferation, differentiation, and vascular remodeling and are also implicated in the pathogenesis of different diseases, including cardiovascular diseases (CVD), diabetes, inflammation, and cancer. 18 members of the KLF family have been distinguished in mammalian cells.5KLF5, also called BTEB2, CKLF, or IKLF in humans, are positioned on chromosome 13q22.1, and encodes a transcription factor protein with 457 amino acids. KLF5 is highly expressed in human skin, esophagus, colon, small intestine, heart, and brain.6 A recent study showed homozygous removal of KLF5 in mice resulted in early embryo death.
In contrast, heterozygous KLF5-knockout led to diminished levels of angiogenesis, decreased neointimal formation, adventitia thickening, cardiac hypertrophy, and interstitial fibrosis in response to angiotensin II injection.7 In this regard, KLF5-knockout in mice cardiomyocytes caused decreased expression of cardiac PPAR1 and its downstream fatty acid metabolism-associated genes, which led to reduced cardiac fatty acid oxidation, increased triglyceride accumulation, abrogating ATP production, and cardiac invalidity. Moreover, KLF5 overexpression notably increased cellular activity and diminished cellular apoptosis in response to myocardial damage.8,9 Recent clinical trial studies revealed that KLF5 is expressed in coronary atherosclerotic plaques, and its SNPs are associated with human hypertension, familial dilated cardiomyopathy (FDM), and basal and resting metabolic rate (BMR) in Japanese, Chinese and Korean populations, respectively.7,10,11 These findings emphasize the critical function of KLF5 in adaptive cardiovascular remodeling and make it causative to screen KLF5 as a candidate gene for CAD vulnerability.
KLF7 (original name: ubiquitous klf) is located on chromosome 2q32 and encodes a protein with 269 amino acids and is generally expressed at a low scale in adult tissue. However, high-level expression is limited to neuronal tissue. It was reported that KLF7 null mice died within two days of birth, as a result extensive neurological disorders.12 Furthermore, it was demonstrated that KLF7 overexpression suppressed adipogenesis and adiponectin gene expression in adipocytes. Also, KLF7 inhibits glucose-induced insulin secretion in pancreatic β-cells and reduces hexokinase 2 expression in skeletal muscle of diabetes patients.13KLF7 genetic polymorphism's role in obesity and diabetes has been proved in Danish and Japanese subjects, respectively.14,15 Therefore, it can be hypothesized that KLF7 may confer susceptibility to CAD via regulating adipocyte function and glucose metabolism.
The mediation of genetic factors in the pathogenesis of CAD is widely acknowledged; however, to date, just a few genes have been determined to increase susceptibility to CAD. Therefore, large-scale SNP genes are needed. Based on the role of KLF5 and KLF7 in adipose and glucose metabolism and the cardiovascular system and a robust correlation between KLF5 and KLF7 variants with metabolic disorders such as diabetes, CADs, body fat, and hypertension, these genes are considered to be candidate genes for conferring susceptibility to CAD.
Therefore, in this clinical trial we aimed to screen for the association between KLF7 (rs2302870) and KLF5 (rs3812852) SNPs with the risk of CAD for the first time in the globe and in the Iranian population.
MethodsStudy patientsThe case-control study comprised 150 patients with CAD (CAD+) and 150 control subjects without CAD (control) through convenient sampling from people undergoing coronary angiography and general checkup in the Tehran Heart Center Hospital, Tehran province, Iran, in 2019. The control group participants were enrolled at their annual health examination. They did not have any history of CAD, chest symptoms or electrocardiogram abnormalities suggesting CAD. The study did not include participants with renal failure, autoimmune diseases, pulmonary obstruction, liver diseases, malignancy, congenital heart disease, and concomitant cardiomyopathy.
The study protocol was approved by the university's local ethics committee and was planned in accordance with the concepts of the Helsinki Declaration (ethics code: 1400.1038). All the patients signed the informed consent forms before enrollment. The subjects underwent a physical examination and completed a question sheet of individual demographic and clinical information, including body weight, medical history, medication, smoking, body mass index (BMI), history of alcohol and substance abuse, hypertension, and other diseases exclusion criteria. Venous whole blood samples collected in EFTA tubes from the participants after overnight fasting and serum were separated by low-speed centrifugation. Blood cells were supplied at −20°C for DNA extraction, and serum was used for the serology study. CAD was detected by observing intense coronary stenosis (≥50%) in at least one of the three main coronary arteries or their main branches via coronary angiography.
Biochemical analysesSerum total cholesterol (TC), triglycerides (TGs), HDL, LDL, and fasting blood sugar (FBS) levels were evaluated using commercially available kits (Pars Azmoon Inc., Tehran, Iran) with an auto-analyzer (Prestige, Japan).
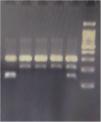
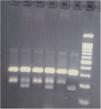
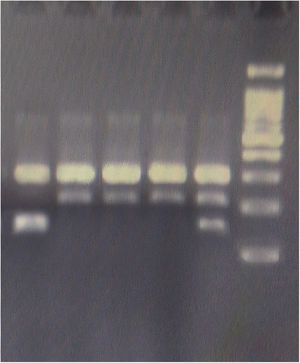
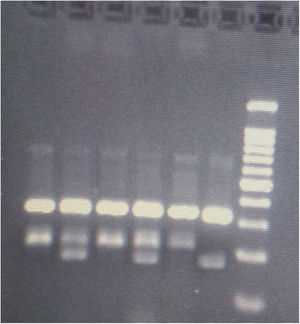
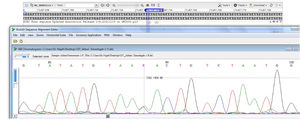
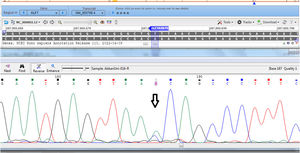
GenotypingGenomic DNA was isolated with a salting-out approach and was checked for quality and quantity by nanodrop spectrophotometry and gel electrophoresis. The KLF5 variant (rs3812852) is located in −1282 bp region within 2000 bp 5ʹ of the transcription initiation location. KLF7 SNP (rs2302870) positioned at putative promoter at intron 2 +35092. Due to the lack of restriction enzymes for these genes, the Tetra Primer ARMS PCR technique was used to detect the single nucleotide variants of the study genes. For this purpose, a pair of outer primers and two allele-specific inner primers were designed for each gene (Table 1). To find the positive control for mutant genotypes, we randomly sequenced 15 samples with Sanger sequencing (ABI) according to the frequency of the mutation before studies. After identifying the mutant samples, we tested different concentrations of primers and DNAs until we reached the desired setup of tetra-ARMS PCR. Genomic DNA was amplified in 15 μl of reaction tube containing 100–200 ng genomic DNA. In this study, for a more accurate junction of primers, the samples were amplified with external primer pairs, then the PCR products were diluted at a ratio of 1–100 and used as a DNA sample for ARMS tetra primer. 0.5 μl of outer F and R primers (10 PM) (Gene fanavaran, Iran), 7 μl master mix containing 200 μmol/L of each dNTP, 1.4 mmol/L MgCl2, and 1 unit of Taq polymerase (Amplicon, Denmark), 1 μl of extracted DNA and 6 μl distilled water were used for first step PCR. For the second step, 0.3 μl of outer F and R primers, 0.4 μl inner F and 0.8 μl inner R primers (10 PM), 7 μl master mix, 1 μl of dilated DNA, and 5.2 μl distilled water were used. PCR cycling plan was 95°C for 5 min (initialization melting) followed by 35 cycles of 95°C for 30 s (denaturing), 57°C for KLF5 and 60 for KLF7 for 30 s (annealing), 72°C for 30 s (extension), with a final extension phase of 5 min at 72°C. The resultant amplicons were evaluated by 2.5% cyber-green gel electrophoresis in 90 V for 40 min. Approximately 30% of cases were randomly evaluated in duplicates for the genotyping quality control, and the concordance rate was 100% (Figures 1–4).
Primers features.
| Gene | Primers sequencing (5′–3′) |
|---|---|
| KLF5 outer F | TGAAAATGAGATCCATACCTTTGATAGA |
| KLF5 outer R | TATGTTGTTATAATGGGAGGGTGAATTT |
| KLF5 inner F (A allele) | TGATGAATAGGCTGTGGTATATGTCAA |
| KLF5 inner R (G allele) | TGTTGACACACAATATACCATTAGACATTC |
| Product size for A allele: 219 | |
| Product size for G allele: 146 | |
| Product size of two outer primers: 308 | |
| KLF7 outer F | CACTCACATCCCTCCCATGAGCACTTAT |
| KLF7 outer R | TCAGAGGAAATGCTCTGACTCTGGGAAC |
| KLF7 inner F (A allele) | CTGGTCAGGCTTGGGAAAGGGCAGT |
| KLF7 inner R (C allele) | GACATCTAGTCTTTCAGGAATGAGTTGAC |
| Product size for A allele: 211 | |
| Product size for C allele: 169 | |
| Product size of two outer primers: 325 | |
All statistical analyses were performed using SPSS (version 19.0) and R (version 3.0.1) software. The Kolmogorov-Smirnov test analyzed the normal distribution of the data. The t-test or analysis of variance was used to analyze the differences between the parametric variables, and the nonparametric variables were analyzed by Mann-Whitney. The Student's t-test or χ2 test compared demographic and clinical data between cases and controls. The chi-square test was utilized to measure the deviation of allelic and genotype frequencies from the Hardy-Weinberg equilibrium expectations. Logistic regression analysis was carried out to estimate the possible effect of the study genotypes on CAD by odds ratios (ORs) and 95% confidence intervals (CI). p values <0.05 were considered to be significant. Data and materials analyzed during the current study are available from the corresponding author upon reasonable request.
ResultsClinical characteristics of the study subjectThere was no statistical difference in the BMI, HDL, TC, and family history of metabolic diseases between the two studied groups. However, older age, being a man, smoking, diabetes, high blood pressure, FBS, LDL, and TG levels were remarkably associated with increased risk of CAD (Table 2).
Clinical characteristics of the study groups.
| CAD+ | Control | p value | |
|---|---|---|---|
| Age | 59.4±9.6 | 53.3±9.8 | <0.05 |
| Gender | Male: 106 (70.6%) | Male: 60 (40%) | <0.01 |
| Female: 44 (29.3%) | Female: 90 (60%) | ||
| Smoking | 31 (20.6%) | 18 (12%) | <0.01 |
| BMI (kg/m2) | 27.5±3.7 | 28.13±4.2 | >0.05 |
| Familial history | 31 (20.6%) | 24 (16%) | >0.05 |
| Hypertension | 95 (63.3%) | 69 (46%) | <0.01 |
| Diabetes | 65 (43.3%) | 17 (11.3%) | <0.001 |
| TG | 179.4±111.7 | 155±86.4 | <0.01 |
| TC | 188.2±50.3 | 179.3±37.4 | >0.05 |
| HDL | 42.6±10.7 | 45.3±10.5 | >0.05 |
| LDL | 126.2±45.12 | 101.2±31.6 | <0.05 |
| FBS | 126.2±52.3 | 101.2±30.3 | <0.01 |
Data are shown as mean±standard deviation. BMI: body mass index; FBS: fasting blood sugar; HDL: high-density lipoprotein; LDL: low-density lipoprotein; TC: total cholesterol; TG: triglycerides.
Univariate logistic regression analysis of KLF7 variants revealed that the distribution of C allele and AC genotypes were statistically higher in the control group compared to CAD+. However, we did not observe any significant association between the KLF5 variants and the risk of CAD, a clinical characteristic of study subjects, according to different genotypes of KLF5 and KLF7 (Tables 3 and 4). Moreover, we analyzed whether the KLF5 and KLF7 SNPs impact diabetes and hypertension risk among CAD+ patients or not, and we found that the distribution of AG genotypes of KLF5 is meaningfully lower in CAD+ with diabetes (D+) patients compared to CAD+ and D− patients (Table 5). However, no significant correlation was found in KLF5 and KLF7 SNPs with the risk of hypertension among CAD+ patients (Table 6).
Genotypes and allelic frequencies in controls and CAD+ patients.
| CAD+ | Control | OR | 95% CI | p value | |
|---|---|---|---|---|---|
| KLF5 | AA 129 (86%) | 125 (83.3%) | 0.54a | 0.218–1.1 | >0.05 |
| AG 20 (13.3%) | 23 (15.3%) | 1.18b | 0.7–2.63 | >0.05 | |
| GG 1 (0.7%) | 2 (1.5%) | 1.41c | 0.23–9.2 | >0.05 | |
| A allele: 92.5% | 91% | >0.05 | |||
| G allele: 7.5% | 9% | 1.38d | 0.6–1.9 | >0.05 | |
| KLF7 | AA 131 (87%) | 122 (81%) | 0.77 | 0.414–1.3 | >0.05 |
| AC 18 (12%) | 27 (18%) | 1.76 | 0.9–3.1 | <0.05 | |
| CC 1 (0.7%) | 1 (0.7%) | 1.81 | 0.27–9.8 | >0.05 | |
| A allele: 93.5% | 90% | >0.05 | |||
| C allele: 6.5% | 10% | 1.89 | 0.8–2.7 | <0.05 | |
Clinical characteristics of study subjects according to different genotypes of KLF5 and KLF7.
| KLF5 (AA) | KLF5 (AG and GG) | p value | KLF7 (AA) | KLF7 (AC and CC) | p value | |
|---|---|---|---|---|---|---|
| Age | 55.3±8.1 | 57.5±7.2 | >0.05 | 57.8±9.2 | 55.4±6.4 | >0.05 |
| BMI (kg/m2) | 26.1±3.7 | 26.9±3.2 | >0.05 | 28.1±3.9 | 27.4±3.4 | >0.05 |
| Hypertension | 135 (53.1%) | 25 (55.9%) | >0.05 | 133 (53.3%) | 27 (56.1%) | >0.05 |
| Diabetes | 69 (27.1%) | 13 (27.7%) | >0.05 | 67 (26.5%) | 14 (28.1%) | >0.05 |
| TG | 163.2±73.2 | 171±59.9 | >0.05 | 170.5±80.8 | 164±63.2 | >0.05 |
| TC | 180.2±37.2 | 186.5±35.1 | >0.05 | 179.4±24.8 | 185.1±23.8 | >0.05 |
| HDL | 45.1±8.3 | 43.3±9.1 | >0.05 | 44.3±10.5 | 43.7±9.3 | >0.05 |
| LDL | 110.5±45.2 | 116.4±40.1 | >0.05 | 109.9±30.3 | 115.1±32.4 | >0.05 |
| FBS | 114.5±43.7 | 110.2±37.3 | >0.05 | 115.6±34.34 | 109.4±30.9 | >0.05 |
Data are shown as mean±standard deviation. BMI: body mass index; FBS: fasting blood sugar; HDL: high-density lipoprotein; LDL: low-density lipoprotein; TC: total cholesterol; TG: triglycerides.
KLF5 and KLF7 genotype distribution in CAD+ patients with (D+) and without (D−) diabetes.
| CAD+ and D+ | CAD+ and D− | OR | 95% CI | p value | |
|---|---|---|---|---|---|
| KLF5 | AA 58 (89.4%) | 73 (85.5%) | 0.59 | 0.216–1.1 | >0.05 |
| AG 6 (10.1%) | 12 (14.5%) | 1.39 | 0.6–2.53 | <0.05 | |
| GG 1 (0.5%) | 0 (0%) | 1.49 | 0.22–9.1 | >0.05 | |
| A allele: 94.4% | 92.8% | >0.05 | |||
| G allele: 5.6% | 7.2% | 1.19 | 0.5–1.8 | >0.05 | |
| KLF7 | AA 57 (87.6%) | 74 (87%) | 0.87 | 0.419–1.4 | >0.05 |
| AC 8 (12.3%) | 10 (11.7%) | 1.86 | 0.1–3.1 | >0.05 | |
| CC 0 (0%) | 1 (1.1%) | 1.91 | 0.29–9.9 | >0.05 | |
| A allele: 93.7% | 93% | >0.05 | |||
| C allele: 6.3% | 7% | 1.99 | 0.9–2.8 | >0.05 | |
KLF5 and KLF7 genotypes distribution in CAD+ patients with (H+) and without (H−) hypertension.
| CAD+ and H+ | CAD+ and H− | OR | 95% CI | p value | |
|---|---|---|---|---|---|
| KLF5 | AA 82 (86.3%) | 46 (83.6%) | 0.65 | 0.22–1.5 | >0.05 |
| AG 12 (12.6%) | 9 (16.4%) | 1.19 | 0.8–2.77 | >0.05 | |
| GG 1 (1%) | 0 (0%) | 1.36 | 0.38–9.1 | >0.05 | |
| A allele: 92.6% | 91.8% | >0.05 | |||
| G allele: 7.3% | 8.2% | 1.49 | 0.7–2 | >0.05 | |
| KLF7 | AA 84 (88.4%) | 47 (87%) | 0.77 | 0.314–1.2 | >0.05 |
| AC 11 (11.5%) | 7 (13%) | 1.26 | 0.7–3.19 | >0.05 | |
| CC 0 (0%) | 1 (1.8%) | 1.21 | 0.27–9.8 | >0.05 | |
| A allele: 94.5% | 91.7% | >0.05 | |||
| C allele: 5.6% | 8.3% | 1.49 | 0.7–2.3 | >0.05 | |
KLF5 was detected in vascular smooth muscle cells of the rabbit aorta for the first time. KLF5 has been determined to be downregulated during development but stimulated in the neointima after damage in the adult aorta to induce cell proliferation and angiogenesis, perhaps via elevating PDGFA expression. Previous investigations have reported that KLF5 can regulate the expression of several target genes such as TGFβ, PPAR1, PDGF-A, and IGF1 in the heart, which have an essential function in cardiac structural remodeling functional adaptation processes.8,16–18 A recent study indicated that KLF5 knockout rats were eliminated in utero before E8.5 and KLF5 +/− heterozygote rats triggered a decrease in neointimal formation, angiogenesis, adventitia thickening, and fibrosis.19
Oishi et al. reported that the KLF5 rs3812852 A/A and A/G genotypes are associated with an increased risk of hypertension in the Japanese population. They determined that people with the G allele have a weaker response to angiotensinogen II (AII) compared to the A allele. They proved that KLF5 expression and activation are increased in response to AII, and its overexpression is associated with an increment of cardiovascular lesions. Therefore, KLF5 A/A and A/G, change the responsiveness of KLF5 to AII, which may lead to hypertension. Also, they revealed a novel signaling and transcription network, including MEF2A and KLF5 as an important regulator of the pathogenesis of cardiovascular diseases such as hypertension. KLF5 and MEF2 are coexpressed in coronary sclerotic lesions. It was identified that ROS are essential for AII-associated activation of KLF5 and KLF5area novel functional molecules in the ROS-p38 MAPK signaling pathway.7
Ruo-Min and coworkers reported KLF5 as a novel disease gene responsible for familial dilated cardiomyopathy (FDC) in Chinese people. They identified heterozygous mutant KLF5 c.1100T>A; p.(Leu367) increased the risk of FDC; moreover has no transcriptional activity. Furthermore, the variants eliminate the synergistic transactivation between KLF5 and NFK-β1another important transcription factor implicated in FDC.10 Jung Ran and a coworker determined mutant homozygote of KLF5 (rs3782933) is a pivotal factor in increasing basal metabolic rate (BMR) in the Korean population. They revealed that people with the TT genotype had markedly higher BMR than the CC genotype. Also, heavier muscle mass, insulin and C-peptide were higher in TT subjects than in CC.11 Yanagi et al. illustrated that KLF5 is expressed in neurons in the human brain's prefrontal cortex and positioned in granular and pyramidal cells in the hippocampus as the main site of glutamatergic neurotransmission. Furthermore, they determined mutation in KLF5 rs3812852 is accompanied by a higher risk of schizophrenia in the Japanese population.20
Based on this research, KLF5 polymorphism contributed to FDC and BMR, and the rs3812852 polymorphism is associated with hypertension (an important risk factor of CAD) and schizophrenia. Therefore, we considered that investigating the possible relationship between rs3812852 polymorphism and CAD for the first time in the world, might be useful to clarify the molecular pathogenesis and genetic susceptibility of CAD. Our results showed that the frequency of the rs3812852 A allele is greater (92.5% vs. 84% and 90%), and the G allele is less (7.5% vs. 16% and 10%) compared to the Japanese population. We failed to find any meaningful difference in KLF5 SNP distribution among CAD+ and the control population. But more analysis revealed that CAD+ patients with AG genotypes have less risk of developing to diabetes in comparison with AA genotypes.
KLF7 has been indicated to block the expression of multiple genes implicated in adipogenesis, including adiponectin, leptin, PPARg, CCAAT, and CEBPa. Previous studies have shown that the expression of Klf7 is suppressed in response to the differentiation of 3T3-L1 adipocytes. Moreover, it has been demonstrated that green tea polyphenol (K)-catechin is able to increase adiponectin expression and glucose uptake into 3T3-L1 adipocytes via downregulation of KLF7. Furthermore, it was determined that KLF7 inhibits glucose-induced insulin secretion in pancreatic β-cells and reduces hexokinase 2 expression in skeletal muscle of T2D patients.13,21 These findings induce the hypothesis that KLF7 can impact cardiovascular function through regulating the expression of the genes involved in glucose metabolism and adipocyte function. Zobl et al. reported no statistical difference in the frequency of the KLF7 rs2302870 and rs7568369 SNPs among T2D and control subjects in Danish people, however the minor A allele of KLF7 rs7568369 protected against obesity, and the frequency of the minor C allele of rs2302870 was remarkably higher in overweight/abdominally obese subjects and abdominally obese individuals compared with lean individuals.15 Kanazawa and a coworker reported that the frequency of heterozygote mutants in KLF7 rs2302870 is statistically lower in T2D patients than in control participants in the Japanese population.14 Based on the research, KLF7 rs2302870 polymorphism contributed to obesity and diabetes, which is an important risk factors for CAD. In this regard, we hypothesize that investigating the possible relationship between rs2302870 polymorphism and CAD for the first time in the world, might be useful to clarify the molecular pathogenesis and genetic susceptibility of CAD. Our research has shown that the frequency of the minor C allele of KLF5 rs2302870 is lower than in Danish and Japanese people (8.5% vs. 10.5%) and meaningfully lower in CAD+ patients than in control participants.
We had some limitations when conducting this study, including sampling during the COVID-19 pandemic, the absence of restriction enzymes for the SNPs to perform the RFLP method and low frequency of some genotypes.
ConclusionIn conclusion, the current study identifies KLF7 gene as a novel candidate for conferring genetic susceptibility to CAD, which provides innovative insights into the molecular pathogenesis of the disease. The exact mechanism is unknown, but it could be speculated that a single nucleotide mutation in the gene caused an alteration its transcriptional activities leading to an increased risk of CAD. We have determined that the KLF5 SNP is unlikely to be implicated in CAD risk. However, in the study population, a mutation in the SNP may have a protective role against the development of diabetes among CAD+ patients. This was the first study to make an association between KLF5 and KLF7 SNP and CAD risk. However, more functional studies are required to clarify the exact correlation of other polymorphisms in KLF5 and KLF7 with the risk of CAD and other metabolic diseases in different recesses.
Learning pointsThe current study identifies KLF7 gene as a novel candidate for conferring genetic susceptibility to CAD, which provides novel insight into the molecular pathogenesis of the disease. A single nucleotide mutation in the gene caused altering its transcriptional activities may lead to an increased risk of CAD. We have determined that the KLF5 SNP is unlikely to be implicated in CAD risk. However, in the study population, a mutation in the SNP may have a protective role against the development of diabetes among CAD+ patients. This may occur as angiotensinogen and NF-KB signal transduction is affected. This was the first study to make an association between KLF5 and KLF7 SNP and CAD risk.
Conflict of interestsAll the authors declare that they have no competing interests.
The authors thank the head and staff of the Tehran Heart Center Hospital, Tehran, Iran, who provided the specimens for the study.