The authors report the clinical and genetic investigation of a family with hypertrophic cardiomyopathy (HCM). The individuals described are three affected first-degree relatives (father, daughter and son), one affected niece and unaffected nephew and niece. Those affected all share a very similar phenotype consisting of asymmetric HCM, with hypertrophy particularly affecting the septum and the anterior wall, and similar electrocardiographic features, including a short PR interval. Case 1 (proband) presented with obstructive HCM and had undergone myectomy and mitral valve replacement. Case 2 (oldest offspring of Case 1) had non-obstructive HCM with exertional angina and NYHA II heart failure (HF) symptoms; she developed non-sustained ventricular tachycardia during follow-up and received a single-chamber ICD for primary prevention of sudden cardiac death. Case 3 (son of case 1) presented with asymptomatic non-obstructive HCM and developed NYHA II HF symptoms during follow-up. Case 4 had non-obstructive HCM, mainly with NYHA II HF symptoms. Testing of the proband for sarcomeric mutations and phenocopies was initially negative. After eight years of clinical follow-up, the suspicion of an undiscovered pathogenic gene mutation shared among the members of this family led us to enroll the proband in a whole-genome sequencing research project, which revealed a heterozygous pathogenic intronic MYBPC3 variant (c.1227-13G>A [rs397515893]), cosegregating with the phenotype.
Os autores descrevem a investigação clínica e genética de uma família com miocardiopatia hipertrófica (MCH). Os casos descritos incluem três parentes de primeiro grau (pai e dois filhos) e uma sobrinha com MCH e dois sobrinhos sem MCH. Todos os casos afetados apresentavam um fenótipo semelhante, que consistia em MCH assimétrica, com hipertrofia envolvendo o septo e a parede anterior e alterações eletrocardiográficas idênticas, incluindo um intervalo PR curto. O caso 1 (índice) apresentava uma MCH obstrutiva e tinha sido previamente submetido a miectomia e substituição valvular mitral. O caso 2 (filha mais velha) apresentou-se com uma MCH não obstrutiva, sintomas de angina e insuficiência cardíaca (IC) classe II de NYHA; desenvolveu taquicardia ventricular não mantida (TVNM) no seguimento e foi submetida a implantação de CDI, em prevenção primária de morte súbita cardíaca. O caso 3 (filho) apresentou-se com MCH não obstrutiva assintomática, tendo desenvolvido queixas de IC classe II de NYHA durante o seguimento. O caso 4 (sobrinha) apresentou-se com MCH não obstrutiva, predominantemente com sintomas de IC clase II NYHA. A pesquisa de mutações sarcoméricas e fenocópias no caso-índice foi, inicialmente, «negativa». Após oito anos em seguimento clínico e perante a suspeita de um novo gene patogénico/nova mutação partilhada entre todos os parentes, incluímos o caso-índice num projecto whole-genome sequencing. Foi diagnosticada uma variante intrónica patogénica, em heterozigotia, do gene MYBPC3 (c.1227-13G>A (rs397515893)), com cossegregação familiar.
Hypertrophic cardiomyopathy (HCM) is a genetic disease of the myocardium transmitted in an autosomal dominant pattern and affecting both genders and all ethnicities,1,2 with a prevalence of 1:500-1:200 in the general population.3 Histologically, it is characterized by myofibrillar disarray and fibrosis, resulting in diastolic dysfunction in virtually all cases.4 In index cases, the diagnosis is established by a maximum wall thickness (MWT) of ≥15 mm by cardiac imaging in the absence of abnormal loading conditions sufficient to explain the hypertrophy.5,6 When the patient is a first-degree relative of a diagnosed HCM patient, a threshold of 13 mm is sufficient to establish the diagnosis.5 HCM is characterized by a wide clinical spectrum, varying from asymptomatic to heart failure or sudden cardiac death (SCD), which can be the first presentation.5,7
The disease is associated with mutations in 11 or more genes encoding contractile myofilament protein components of the sarcomere or proteins of the adjacent Z-disc.8 More than 1400 mutations (largely missense) have been identified, most of which are unique to individual families.5 Among genotype-positive patients, 70% have mutations in beta-myosin heavy chain (MYH7) and myosin-binding protein C (MYBPC3) genes.5 Genetic testing confirms the diagnosis and enables close follow-up and monitoring of genotype-positive relatives before they develop hypertrophy.5,9,10
The DNA resequencing methods used for clinical purposes, Sanger sequencing or more recently next-generation sequencing of targeted panels, only screen exons and immediately contiguous exon-intron boundaries. A sarcomeric mutation is found in approximately 40-50% of probands.11 Deeper intronic variants or new undiscovered genes are two possible explanations for this incomplete yield, as we hypothesised in this family, studied by whole-genome sequencing after years of follow-up.
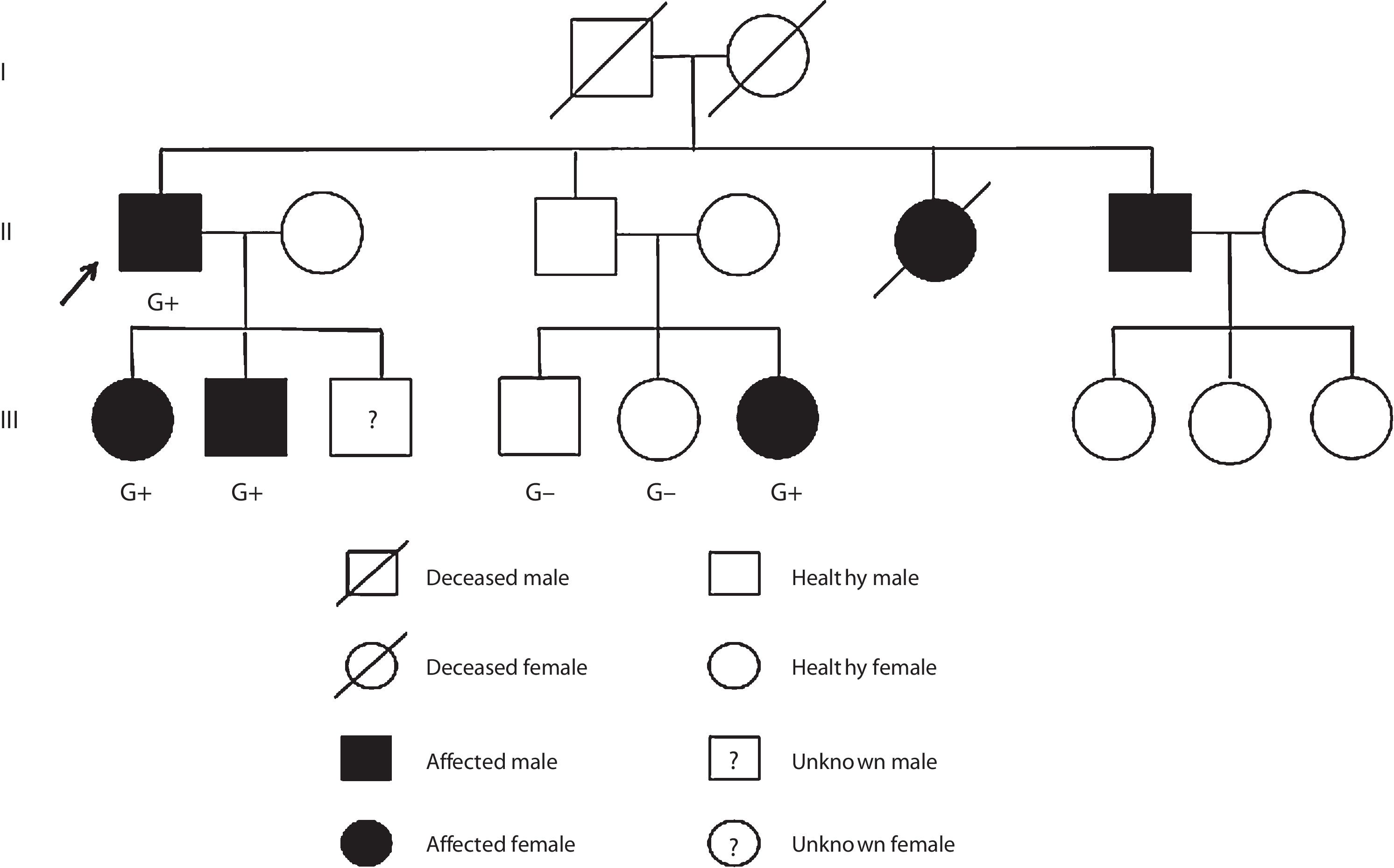
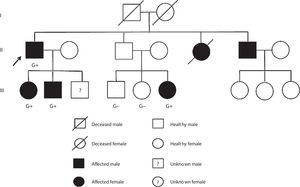
Case reportCase 1 (II-1) (proband)A 76-year-old man with previous history of hypertension, dyslipidemia, and paroxysmal atrial fibrillation was diagnosed with obstructive HCM at the age of 52 years. He was known to have a brother diagnosed with HCM at the age of 55 years (II-6) and a sister who died suddenly of unknown cause at the age of 49 years (II-5). The family pedigree is shown in Figure 1.
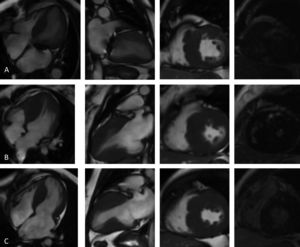
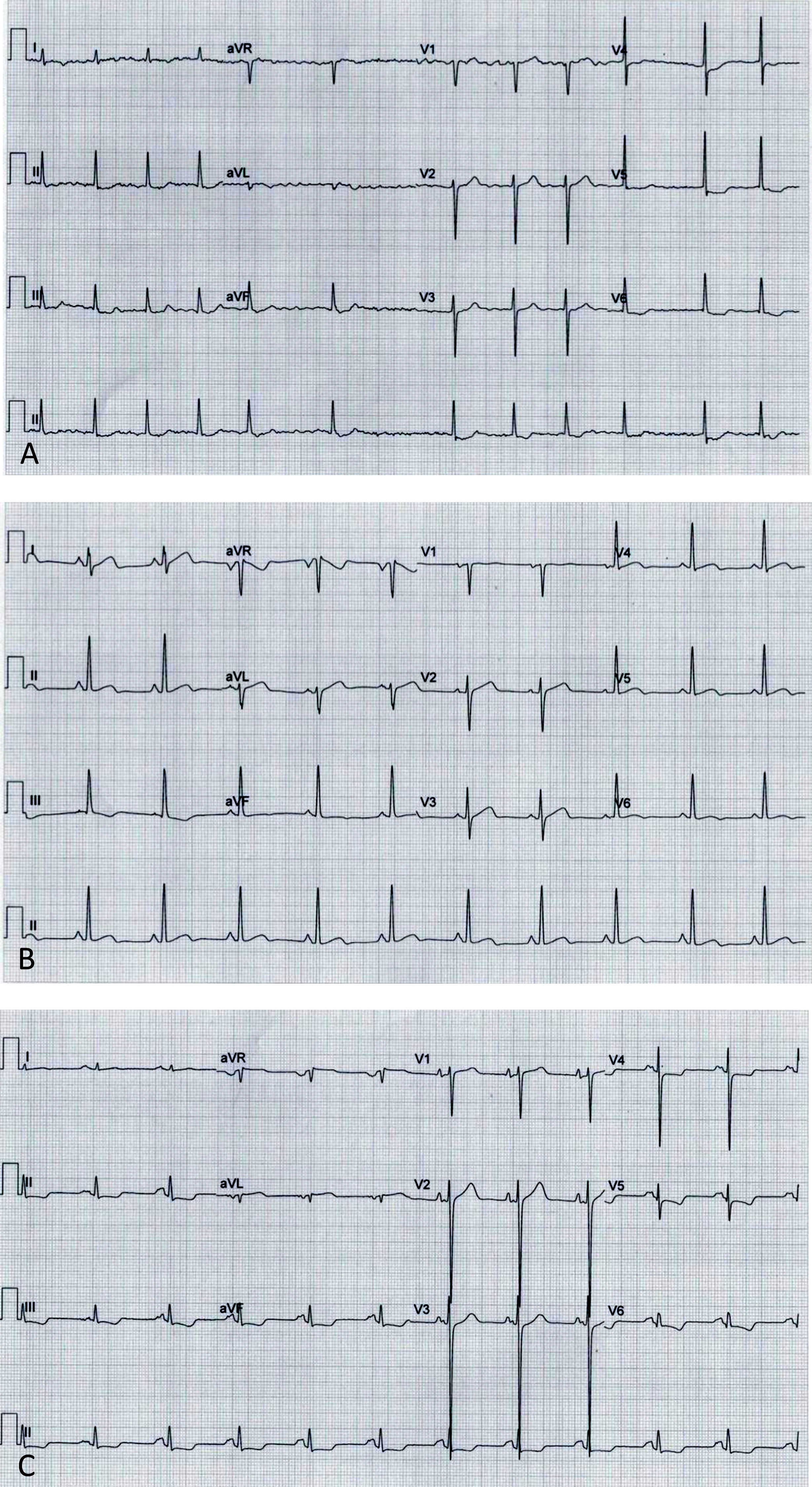
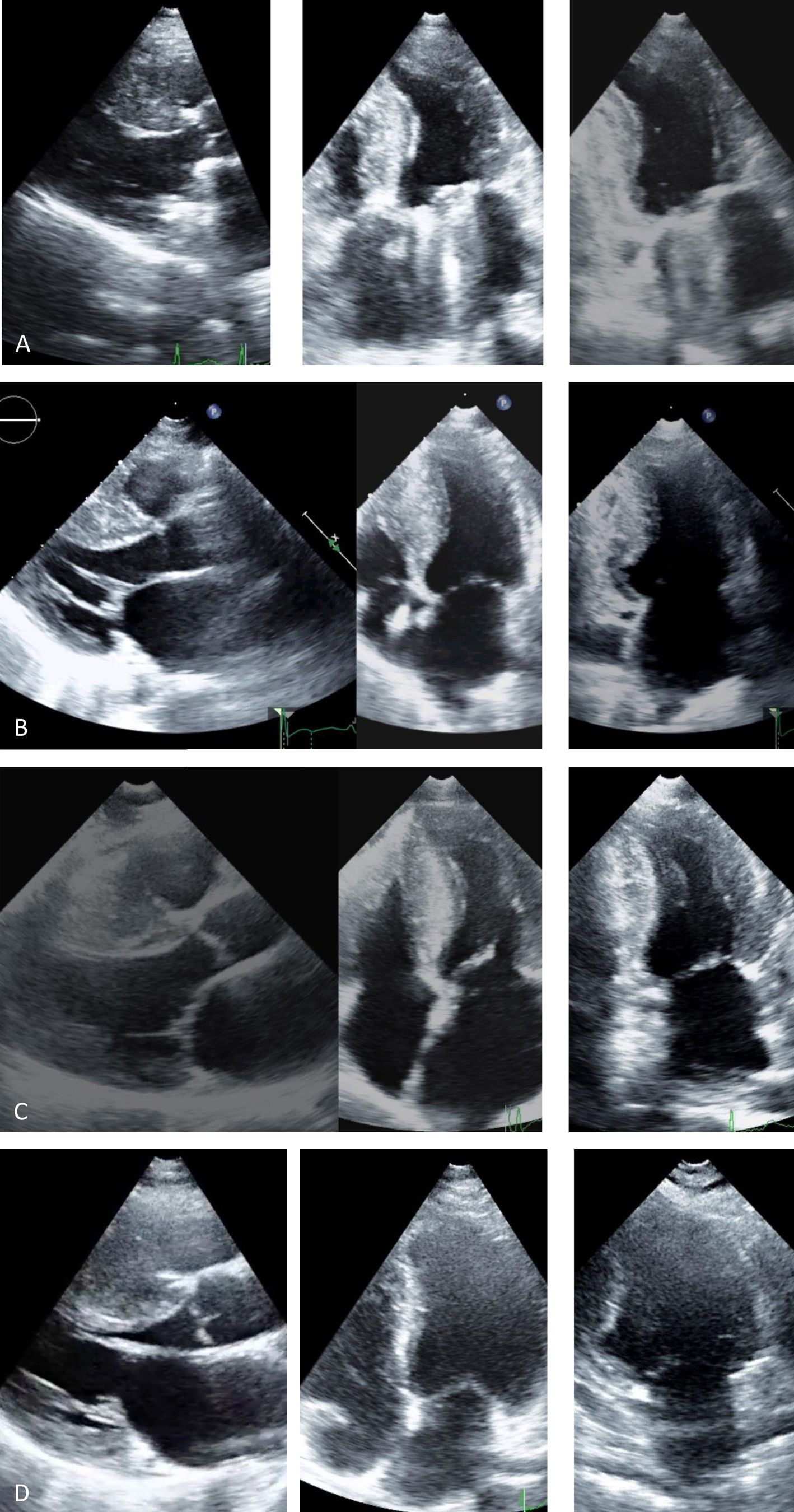
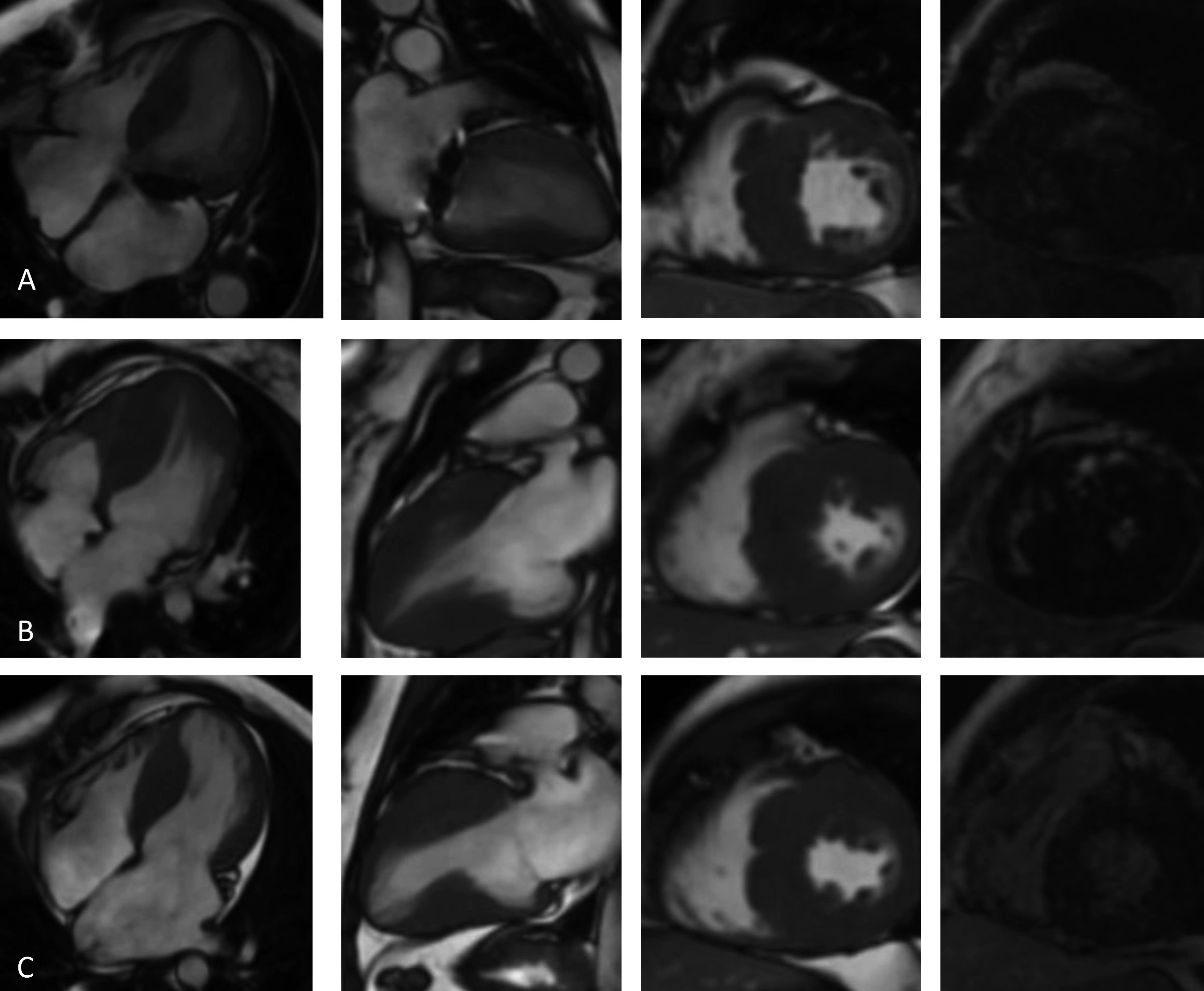
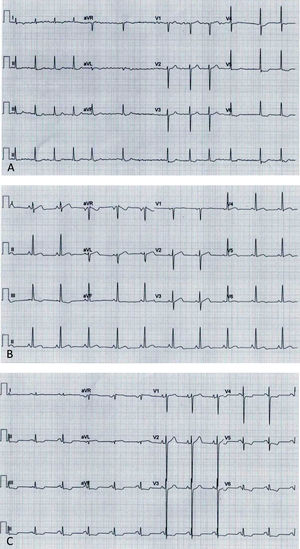
Before follow-up in our clinic, he had undergone myectomy and mitral valve replacement for left ventricular outflow tract (LVOT) obstruction refractory to medical therapy at the age of 60 years. He has been in New York Heart Association (NYHA) class II-III heart failure (HF) ever since, treated with oral anticoagulants, angiotensin-converting enzyme inhibitors, spironolactone and beta-blockers. After four years of uneventful follow-up, he had an embolic stroke. His ECGs showed atrial fibrillation, ST-segment depression and biphasic T-waves in V4-V6, III and aVF (Figure 2A). The transthoracic echocardiograms showed severe septal hypertrophy (interventricular septum 19 mm, inferolateral wall 13 mm), left ventricular ejection fraction (LVEF) 55%, increased filling pressures (E/e’ ratio 31) with no exercise-induced intraventricular gradient, and a normally functioning mechanical mitral valve (Figure 3A). Some of the 24-hour Holter recordings during follow-up showed non-sustained ventricular tachycardia (NSVT) as the only American College of Cardiology Foundation/American Heart Association risk factor,12 and his last estimated European Society of Cardiology SCD risk score5,13 was 5.0% at five years. Cardiac magnetic resonance (CMR) showed asymmetric HCM (MWT 24 mm), normal systolic function (LVEF 61%) and focal late gadolinium enhancement (LGE) in the hypertrophied segments (Figure 4A).
Electrocardiograms: (A, Case 1): atrial fibrillation, ST-segment depression and biphasic T-waves in V4-V6, III and aVF; (B, Case 2): sinus rhythm, short PR interval, T-wave inversion in III, flat T-wave in aVF, biphasic T-wave in II, V4-V6; (C, Case 3): sinus rhythm, short PR interval, electrocardiographic criteria for left ventricular hypertrophy, ST-segment depression and T-wave inversion in V4-V6, II, III and aVF.
Transthoracic echocardiogram, parasternal long-axis, 4-chamber and 2-chamber views: (A, Case 1): asymmetric hypertrophic cardiomyopathy (HCM) (maximum wall thickness [MWT] 16 mm) and normal prosthetic mitral function; (B, Case 2): asymmetric HCM (MWT 22 mm); (C, Case 3): asymmetric HCM (MWT 21 mm); (D, Case 4): asymmetric HCM (MWT 22 mm).
Cardiac magnetic resonance imaging (4-chamber, 2-chamber and short-axis cine and late gadolinium enhancement (LGE) views): (A, Case 1): asymmetric hypertrophic cardiomyopathy (HCM) with late gadolinium enhancement (LGE) in the hypertrophied segments; (B, Case 2): asymmetric HCM with LGE in the hinge points; (C, Case 3): asymmetric HCM with LGE in the hinge points.
The proband underwent clinical genetic testing in 2009, which was reported negative for sarcomeric mutations in MYH7, MYBPC3, TNNT2, TNNI3, MYL2, MYL3, ACTC1, TMP1, and CSRP3, extended three years later to PRKAG2, GLA and LAMP2, also negative. The search for mutations in each of these genes, in an accredited clinical laboratory, was performed for the entirety of the coding region, including exon-intron boundaries (canonical splice sites), using polymerase chain reaction and direct Sanger sequencing. This method has 99% sensitivity in detecting nucleotide substitutions and short insertion-deletions (indels).
In 2017, DNA extracted from the proband's blood was sent to Novogene Corporation for whole-genome sequencing (the rationale for this decision is explained in the discussion). A capture library from New England Biolabs was used for whole-genome amplification and sequencing was carried out on an Illumina HiSeq X Ten platform, with 150 bp paired-end reads and a mean read depth of 50. Reads were pre-processed with Trim Galore version 0.4.4 to remove sequencing adapters and to filter the sequences with a minimum Phred quality score of 20. Most of the reads (∼98.8%) were kept for downstream alignment against the hg19 reference genome using Burrows-Wheeler Aligner (BWA) mem version 0.7.15. The Genome Analysis Toolkit version 4.0.1.2 (GATK4) was applied to mark duplicates and to calculate genome coverage. Variant calling was performed with Strelka2 version 2.8.4 at a mean genome coverage of 40×. After an initial filtering step, 4 862 291 variants (both single-nucleotide polymorphisms and small indels) were obtained. Further analysis revealed a heterozygous intronic variant, c.1227-13G>A (rs397515893), in the MYBPC3 gene (intron 14), classified as likely pathogenic.
The proband's relatives were then genotyped for this variant by Sanger sequencing. As shown in Figure 1, the variant was present in all four affected members and not present in the three non-affected members who were tested.
Case 2 (III-1)The proband's oldest daughter (Figure 1) was referred for clinical screening at the age of 38 years. At that time, she was in NYHA class II and had had previous episodes of exertional angina. The physical examination revealed a harsh systolic murmur at the lower left sternal border. The ECG showed sinus rhythm, short PR interval, T-wave inversion in DIII, flat T wave in aVF and biphasic T wave in DII and V4-V6 (Figure 2B). The transthoracic echocardiogram confirmed the diagnosis, showing asymmetric HCM involving the septum and anterior wall (MWT 22 mm), with no systolic anterior motion (SAM) of the mitral valve or intraventricular gradient at rest or with Valsalva maneuver or exercise (Figure 3B). As well as confirming the echocardiographic findings, CMR revealed mid-cavity obliteration and LGE matching the hypertrophic segments and the left ventricular-right ventricular (LV-RV) hinge points (Figure 4B). She was started on a beta-blocker. At that time, she had no risk factors for sudden cardiac death. Two years later she developed short episodes of palpitations and 24-hour Holter monitoring showed non-sustained ventricular tachycardia. She had an estimated SCD risk of 5.9% at five years (age: 42 years, MWT: 22 mm, left atrial size: 48 mm, maximum LVOT gradient: 2 mmHg; family history of SCD: no, non-sustained ventricular tachycardia: yes, syncope: no) and received a single-chamber implantable cardioverter-defibrillator (ICD) for primary prevention of SCD. Three years later she was in NYHA class III HF. The transthoracic echocardiogram showed a restrictive diastolic filling pattern (E/e’ ratio 18), diminished longitudinal systolic function (septal S’ 3 cm/s and lateral S’ 6 cm/s; global longitudinal strain -11%) and hypokinesia of the hypertrophic segments despite normal ejection fraction; she also had incomplete SAM but no significant mitral regurgitation. She never developed significant intraventricular gradients during follow-up, including on treadmill exercise echocardiography. Under spironolactone, her functional capacity improved and the next transthoracic echocardiogram also showed an improvement in LV filling pressures (E/e’ ratio 9). She has had no malignant arrhythmia since.
Case 3 (III-2)The proband's middle son (Figure 1) was first studied in the setting of familial screening at 35 years of age. Although he was asymptomatic, his transthoracic echocardiogram confirmed the diagnosis of asymmetric HCM, with hypertrophy particularly involving the septum (MWT 21 mm) and the anterior and inferior walls, mild mitral regurgitation and no other relevant findings (Figure 3C). He underwent treadmill exercise echocardiography, which showed no exercise-induced obstruction. The baseline ECG was unremarkable except for evidence of left atrial abnormality. The 24-hour Holter ECG findings were also unremarkable. CMR showed asymmetric HCM (MWT 24 mm), with normal systolic function (LVEF 75%) and focal LGE in the LV-RV hinge points (Figure 4C). After two years, he developed NYHA class II HF symptoms and exercise-induced dizziness. The ECG at that time showed sinus rhythm, short PR interval, slow R-wave progression in the precordial leads and T-wave inversion in the inferior and lateral leads. The transthoracic echocardiogram revealed increased LV filling pressures (mean E/e’ ratio 16). He was started on spironolactone and remained in NYHA class II HF. Three years later, although clinically stable, he developed electrocardiographic criteria for left ventricular hypertrophy (LVH), as well as a short-PR interval, deep S waves in the right precordial leads, ST-segment depression and T-wave inversion in V4-V6 and the inferior leads (Figure 2C). The transthoracic echocardiogram was similar to the previous one. During follow-up, 24-hour Holter monitoring showed no relevant arrhythmia and the echocardiogram revealed no significant intraventricular obstruction, including on treadmill stress testing. His estimated SCD at last clinical follow-up was 3.1% at five years (age: 39 years, MWT: 25 mm, left atrial size: 50 mm, maximum LVOT gradient: 2 mmHg; family history of SCD: no, non-sustained ventricular tachycardia: no, syncope: no).
Case 4 (III-6)This niece of the proband (Figure 1) was mostly followed in another center abroad. She was diagnosed with HCM at the age of 24 and at early presentation she was in NYHA class II HF. At that time, 24-hour Holter monitoring showed no NSVT. She was referred to our clinic during her second pregnancy (11 years after diagnosis). The ECG showed sinus rhythm, short PR interval, electrocardiographic criteria for LVH, QS complexes in DIII and aVF and T-wave inversion in V2-V5. Her transthoracic echocardiogram (38th week of pregnancy) showed asymmetric HCM affecting the septum (MWT 22 mm), SAM of the mitral valve but no significant gradient, normal systolic function and increased filling pressures (E/e’ ratio 21) (Figure 3D). The transthoracic echocardiogram showed normal filling pressures after delivery (E/e’ ratio 6).
Cases 5 and 6 (III-4 and III-5)Cases 5 and 6 were niece and nephew of the proband (Figure 1), enrolled in the clinical screening. Their electrocardiograms and echocardiograms showed no signs of HCM.
DiscussionWe describe a family with HCM considered genotype-negative for eight years of follow-up. The availability of whole-genome sequencing proved to be crucial to obtaining a genetic diagnosis of a pathogenic intronic (splice-site) variant.
The phenotype in this family was relatively homogeneous, with asymmetric HCM, mainly HF symptoms and short PR interval in three of the family members (the proband presented with atrial fibrillation and no previous ECG was available to assess his PR interval). Disease progression was characterized by worsening HF and increased filling pressures. However, the initial screening for sarcomeric mutations was negative, and so the genetic cause could not be found. We first thought this could be due to the presence of a phenocopy. The evidence of a red flag (short PR interval) in three affected relatives increased this suspicion.14 Short PR interval can occur in Anderson-Fabry disease (AFD), Danon disease and PRKAG2 cardiomyopathy. The latter is a rare autosomal dominant disease characterized by heart-specific non-lysosomal glycogenosis, usually diagnosed in late adolescence and characterized by an HCM-like phenotype and evidence of short PR interval and/or pre-excitation, frequently progressing to high-grade conduction system disease.15 The presence of male-to-male transmission excluded the possibility of Danon disease or AFD. However, the proband's son's phenotype status was not known when we initially started following his father, and genetic testing was therefore extended to include these three phenocopies; again, no causal mutation was found.
The main sarcomeric and sarcomere-associated genes and phenocopies had been tested in the proband and these are still the most robustly established causal genes in HCM.16,17 There have recently been increasingly frequent reports that expansion of gene panels to include further cardiomyopathy candidate genes has a nearly negligible effect on testing yield, while generating a higher number of variants of unknown significance and of challenging interpretation.18–21 As such, given a relatively phenotypically homogeneous familial HCM with an undiscovered mutation shared between the four affected cases and with affected and non-affected relatives available to test, we hypothesised that the use of whole-exome sequencing (WES) or whole-genome sequencing (WGS) could lead to the discovery of a new causal gene. Conventional genetic testing only screens exons and immediate intron-exon boundaries, while variants located further within intronic regions tend to be missed.16 However, recent studies have shown that mutations in intronic regions may play a role in HCM patients.22,23 One of these studies22 reported four likely pathogenic deep-intronic variants in MYBPC3 in a small cohort of 58 probands, and we recently published a preliminary quantitative analysis of non-coding variants which suggested that deep-intronic variation contributes to the HCM phenotype.22 Indeed, the presence of a previously undetected pathogenic non-coding variant could have explained the sarcomeric-negative status of this family. Furthermore, the use of next-generation sequencing – WES or WGS – could additionally enable a search for large indels and copy-number variants, through the use of well-established read-depth methods.24
For these reasons, we opted to include this family in a whole-genome sequencing research project, starting with the index case.
Whole-genome sequencing in the proband revealed a heterozygous intronic (splice-site) MYBPC3 variant considered very likely pathogenic (c.1227-13G>A [rs397515893]). This mutation had previously been described in HCM, as documented in the ClinVar database.25 This sequence change falls in intron 14 of the MYBPC3 mRNA. It does not directly change the encoded amino acid sequence of cardiac myosin-binding protein C, but causes a splicing defect that results in a frameshift and is predicted to result in a premature stop codon leading to an absent or truncated protein (PMID: 12110947). We have confirmed cosegregation in the family.
ConclusionWhole-genome sequencing is still not clinically available or indicated for the majority of HCM patients. However, its role in cases of clearly familial HCM for which genetic testing was negative is beginning to be recognized. In the case presented, it enabled the diagnosis of a causal intronic (splice-site) variant shared by the affected family members and previously described as likely pathogenic.
FundingThis work was supported by projects funded by the European Regional Development Fund (FEDER) through POR Lisboa 2020 - Programa Operacional Regional de Lisboa PORTUGAL 2020, and Fundação para a Ciência e a Tecnologia (LISBOA-01-0247-FEDER-024136, and PAC-PRECISE-LISBOA-01-0145-FEDER-016394).
Conflicts of interestThe authors have no conflicts of interest to declare.


![Transthoracic echocardiogram, parasternal long-axis, 4-chamber and 2-chamber views: (A, Case 1): asymmetric hypertrophic cardiomyopathy (HCM) (maximum wall thickness [MWT] 16 mm) and normal prosthetic mitral function; (B, Case 2): asymmetric HCM (MWT 22 mm); (C, Case 3): asymmetric HCM (MWT 21 mm); (D, Case 4): asymmetric HCM (MWT 22 mm). Transthoracic echocardiogram, parasternal long-axis, 4-chamber and 2-chamber views: (A, Case 1): asymmetric hypertrophic cardiomyopathy (HCM) (maximum wall thickness [MWT] 16 mm) and normal prosthetic mitral function; (B, Case 2): asymmetric HCM (MWT 22 mm); (C, Case 3): asymmetric HCM (MWT 21 mm); (D, Case 4): asymmetric HCM (MWT 22 mm).](https://static.elsevier.es/multimedia/21742049/0000003900000004/v1_202010170736/S2174204920301744/v1_202010170736/en/main.assets/thumbnail/gr3.jpeg?xkr=ue/ImdikoIMrsJoerZ+w9znTMwFdb/TnkS0koegILxs=)