Clopidogrel requires oxidation dependent on the cytochrome P450 enzyme 2C19 (CYP2C19) to form its active metabolite. The importance of loss-of-function alleles (particularly CYP2C19*2, 681G>A) in poor platelet response to clopidogrel is well recognized.
ObjectiveTo investigate the prevalence and prognostic impact of the CYP2C19*2 allele in a local acute coronary syndrome (ACS) population.
MethodsWe performed a prospective, longitudinal study of 95 patients admitted for an ACS between March and October 2009 to a single coronary care unit. Patients aged under 75 who survived hospital stay and for whom clopidogrel was prescribed were included. At discharge, CYP2C19 was genotyped using a commercially available kit. Patients were divided into two groups: Group A (non-carriers, normal metabolizers, CYP2C19*1/*1), n=69; and Group B (carriers, slow metabolizers, CYP2C19*2/*1 or *2/*2), n=26. The primary endpoint was a combined outcome of cardiovascular death, non-fatal myocardial infarction or re-admission for unstable angina; median follow-up was 136.0 (79.0–188.0) days.
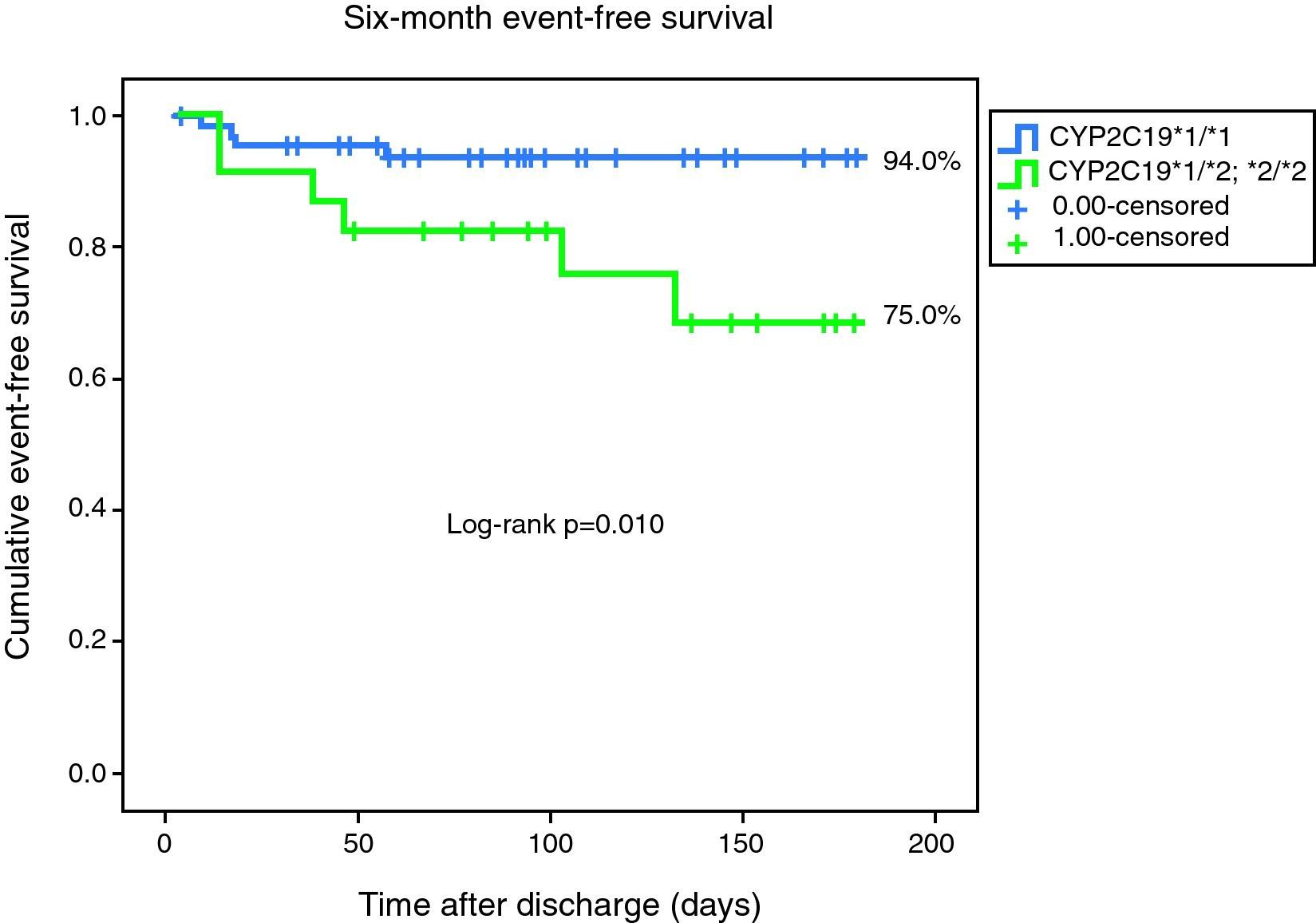
ResultsThe median age of the population was 62.0 (51.0–68.0) years, and 83.2% were male. The CYP2C19*2 (A) allele had a frequency of 14.2%. There were no differences between the groups with respect to demographic data or history of cardiovascular disease. Coronary anatomy, left ventricular ejection fraction and renal function were also similar. The groups were also homogenous with respect to GRACE risk score (118.0 (95.0–136.5) vs. 115.0 (96.0–133.0), p=0.68), medical treatment and percutaneous revascularization during hospital stay. Event-free survival was higher for Group A (94.0% vs. 75.0%, log-rank p=0.010). Three readmissions for MI were documented, all in the slow metabolizers group.
ConclusionIn our ACS population, the CYP2C19*2 allele was a medium-term prognostic marker.
Polimorfismos no complexo enzimático P450, como CYP2C19*2, 681G>A parecem estar na base de fraca resposta ao clopidogrel.
ObjectivoCaracterizar a prevalência e o impacto prognóstico do alelo CYP2C19*2(A) numa população local de doentes com síndrome coronária aguda (SCA).
População e métodosEstudo longitudinal, prospectivo, de 95 doentes admitidos por SCA entre Março e Outubro de 2009. Foram incluídos doentes com idade inferior a 75 anos, que sobreviveram à hospitalização, e que tiveram alta medicados com clopidogrel. Foi determinado o genótipo do alelo CYP2C19 no momento da alta hospitalar. Foram criados dois grupos; grupo A (metabolizadores normais, CYP2C19 *1/*1) n=69 e grupo B (metabolizadores lentos, CYP2C19 *2/*1 ou *2/*2) n=26. No seguimento foi analisado um resultado combinado de morte cardiovascular, enfarte agudo do miocárdio não fatal, e readmissão por angina instável (tempo mediano de seguimento: 136 (79–188) dias.
ResultadosA idade mediana da população era de 62,0 (51,0–68,0) anos, e 83,2% eram do género masculino. O alelo CYP2C19*2(A) teve uma frequência de 14,2%. Não existiram diferenças entres os grupos relativamente a dados demográficos, e antecedentes cardiovasculares. A anatomia coronária, a fracção de ejecção ventricular esquerda e a função renal eram semelhantes. Os grupos eram também homogéneos relativamente ao score GRACE (118,0 (95,0–136,5) versus 115,0 (96,0–133,0), p=0,68), tratamento médico e taxa de revascularização percutânea durante hospitalização. No final do seguimento clínico a sobrevida livre de eventos foi superior para o grupo A (94,0% versus 75,0%, log rank p=0,010). Foram documentadas três readmissões por enfarte agudo do miocárdio, todas no grupo B.
ConclusõesNa população referida, o alelo CYP2C19*2 (A) foi um marcador de prognóstico a médio prazo.
Dual antiplatelet therapy with aspirin and clopidogrel appears to offer synergistic benefit in preventing thrombus formation, but up to 15% of high-risk acute coronary syndrome (ACS) patients continue to suffer from ischemic events.1 A recent meta-analysis found an overall prevalence of 21% of laboratory defined clopidogrel non-responsiveness.2 The mechanisms leading to poor response to clopidogrel and therefore to high platelet reactivity (HPR) have not been fully elucidated and are probably multifactorial. Compliance, cellular, environmental, genetic and clinical factors such as obesity, diabetes mellitus, nature of coronary injury and inflammation are known to contribute to variable antiplatelet drug response.3
Response to clopidogrel may be influenced by pharmacokinetic variables such as intestinal absorption, metabolic activation in the liver, and biologic activity.4 Clopidogrel is a prodrug and requires oxidation that is mainly dependent on the cytochrome P450 enzymes 2C19 (CYP2C19) and to a lesser extent the isoenzymes CYP2C9, 3A4, 3A5 and 2B6.5 Around 25 genetic variants in CYP2C19 have been found (www.cypalleles.ki.se). The presence of the loss-of-function 681G>A polymorphism (*2), a single-nucleotide polymorphism (SNP) (rs4244285) in exon 5 mapped to the long arm of chromosome 10 (10q24.1–q24.3),6 is associated with clopidogrel responsiveness in healthy individuals7 and in patients with coronary artery disease.8,9
Carriers of a reduced-function CYP2C19 genomic variant treated with clopidogrel for an ACS or by percutaneous coronary intervention (PCI) probably have a worse ischemic outcome, as was reported in a recent meta-analysis that included 9685 patients.10 By contrast, data from 2208 patients in FAST-MI (French Registry of Acute ST-Elevation and Non-ST-Elevation Myocardial Infarction) did not identify carriers of a single loss-of-function allele in CYP2C19 with a worse ischemic endpoint, although a significantly higher event rate was noted for carriers of any two or more loss-of-function alleles.4 A negative result was noted in the CHARISMA (Clopidogrel for High Atherothrombotic Risk and Ischemic Stabilization, Management and Avoidance) genomic analysis, as no relationship was seen between CYP2C19*2 heterozygotes and ischemic outcomes, although CYP2C19*2 homozygotes had a worse prognosis.11
In this context, we decided to evaluate the frequency and medium-term prognostic impact of the CYP2C19*2 allele in a local single-center Portuguese ACS cohort.
MethodsStudy design and eligibilityWe performed a prospective longitudinal study of 95 consecutive patients admitted for an ACS who survived hospital stay between April and October 2009.
The local ethics committee approved the research protocol and informed consent was obtained.
To be eligible, patients had to be less than 75 years old and be discharged on clopidogrel therapy (75mg/day).
Patients were excluded if they were immediately referred for surgery, or if they were included in either of two clinical trials related to anti-platelet therapy – TRA*CER (Trial to Assess the Effects of SCH 530348 in Preventing Heart Attack and Stroke in Patients With Acute Coronary Syndrome12 – ClinicalTrials.gov identifier: NCT00527943) and TRILOGY ACS (A Comparison of Prasugrel and Clopidogrel in Acute Coronary Syndrome Subjects – ClinicalTrials.gov identifier: NCT00699998).
ST-elevation myocardial infarction was defined as ischemic chest pain associated with new-onset ST elevation in the ECG greater than 1mm in at least two contiguous leads.
Non-ST elevation myocardial infarction was defined as ischemic chest pain lasting more than five minutes, and positive cardiac biomarkers (troponin I) with or without ischemic ECG changes (ST depression or T-wave inversion).
Unstable angina was defined as new onset angina (at least CCS class III), progressive angina, or angina at rest, with or without ischemic ECG changes.
The clopidogrel loading dose was determined by the cardiologist in the emergency department, and the subsequent daily and discharge dosage was 75mg per day. The aspirin loading dose was determined similarly, with a subsequent and discharge dosage of 100mg per day.
Platelet function tests were performed at discharge. Clopidogrel responsiveness was tested using multiple electrode aggregometry (MEA), on a Multiplate analyzer®.13 MEA was assessed in whole blood; ADP was used as agonist and the results were expressed in arbitrary units (U).14
Baseline and follow-up dataStandardized records at admission included demographic, clinical, electrical and laboratory data. Medical therapy, catheterization data, in-hospital events and discharge medication were also recorded.
Median clinical follow-up was 136.0 (79.0–188.0) days after hospital discharge. The information was collected by telephone, from hospital records or at the outpatient clinic. The primary endpoint analyzed was the combined outcome of cardiovascular death, non-fatal myocardial infarction or re-admission for unstable angina.
GenotypingGenomic DNA was extracted at hospital discharge from peripheral blood leukocytes.
CYP2C19*1 (wild type) and CYP2C19*2 (681G>A), two SNPs in the CYPC2C19 gene, were genotyped using a commercial kit from Seegene®.
The technique employed is based on dual priming oligonucleotide (DPO) polymerase chain reaction (PCR). DPO PCR consists of two separate priming regions joined by a polydeoxyinosine linker. This linker is not involved in priming but rather in delineating the boundary between the two parts of the primer. In the first priming reaction the longer 5′ segment binds to the template DNA, initiating stable annealing. In the second priming reaction the short 3′ segment selectively binds to a target site and determines a target-specific extension, acting as a determiner. Conventional primers have a single priming region and extension may proceed even in the presence of mismatches between a primer and a template. The new methodology used is a fundamental tool for blocking extension of non-specifically primed templates, generating consistently high PCR specificity, as previously described for CYP2C19 SNPs.15
A commercial genomic DNA extraction kit was used in order to obtain 60–100ng of isolated DNA for PCR. Amplification was performed in a final volume of 20μl in accordance with the manufacturer's instructions. It contained 4μl of DNA solution, 3μl MOP solution, 10μl 2× Multiplex Master Mix (Seegene, Seoul, Korea) and 3μl of template.
The reaction started after an initial preheating step at 94°C for fifteen minutes, followed by 35 amplification cycles in the thermal cycler under the following conditions: denaturation at 94°C for 30s, annealing at 63°C for 30s, and extension at 72° C for 30s. Amplification was completed with a final extension step at 72°C for 5min. PCR fluorescence yield for the two different dyes was measured to obtain the allelic discrimination plot and to identify individual genotypes and was presented on a 2-dimensional graph.
Due to the low number of CYP2C19*2 homozygotes, most of the analysis was based on the comparison of CYP2C19*2 (*2/*1 or *2/*2) carriers and wild type homozygotes (*1/*1). Patients were divided into two groups: Group A (non-carriers), n=69; and Group B (carriers), n=26.
Statistical analysisContinuous data are presented as median and interquartile range and groups were compared with the Mann–Whitney test. Categorical variables are reported as frequencies and percentages, and the chi-square test or Fisher's exact test was used as appropriate.
Deviation from Hardy–Weinberg equilibrium in the CYP2C19 polymorphism was tested with the chi-square test.
Cumulative survival curves were constructed by the Kaplan–Meier method and groups were compared with the log-rank test. The observational period started at hospital discharge.
Multivariate Cox regression analysis was performed for the primary endpoint. Variables that were significant at the bivariate level (with a p value below 0.10) and that had clinical relevance were included in the model.
All statistical tests were two-tailed and a p value less than 0.05 was deemed significant. The analysis was performed with SPSS (Statistical Package for the Social Sciences) version 15, SPSS Inc., Chicago, IL.
ResultsThe median age of the population was 62.0 (51.0–68.0) years with a predominance of males (83.2%). All were Caucasian.
Of the 95 patients included, 69 (72.6%) were wild-type homozygous (*1/*1), 25 (26.3%) were heterozygous with respect to the *2 allelic variant (*1/*2), and one (1.1%) was homozygous. The allelic frequency of the CYP2C19*2 (A) allele was 14.2%, and was 85.8% for CYP2C19*1 (G). No deviation was noted from the expected proportions of genotypes in the population predicted by Hardy–Weinberg equilibrium for polymorphisms (p=0.44).
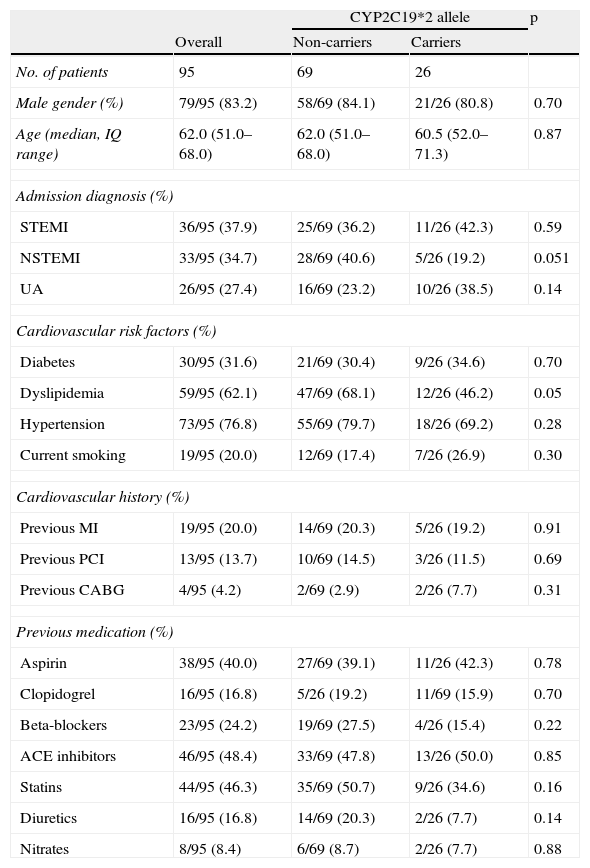
There were no significant differences in terms of demographics, admission diagnosis, cardiovascular history or cardiovascular risk factors, with the exception of previous dyslipidemia, which was more prevalent in the non-carrier group (Table 1).
Baseline characteristics.
| CYP2C19*2 allele | p | |||
| Overall | Non-carriers | Carriers | ||
| No. of patients | 95 | 69 | 26 | |
| Male gender (%) | 79/95 (83.2) | 58/69 (84.1) | 21/26 (80.8) | 0.70 |
| Age (median, IQ range) | 62.0 (51.0–68.0) | 62.0 (51.0–68.0) | 60.5 (52.0–71.3) | 0.87 |
| Admission diagnosis (%) | ||||
| STEMI | 36/95 (37.9) | 25/69 (36.2) | 11/26 (42.3) | 0.59 |
| NSTEMI | 33/95 (34.7) | 28/69 (40.6) | 5/26 (19.2) | 0.051 |
| UA | 26/95 (27.4) | 16/69 (23.2) | 10/26 (38.5) | 0.14 |
| Cardiovascular risk factors (%) | ||||
| Diabetes | 30/95 (31.6) | 21/69 (30.4) | 9/26 (34.6) | 0.70 |
| Dyslipidemia | 59/95 (62.1) | 47/69 (68.1) | 12/26 (46.2) | 0.05 |
| Hypertension | 73/95 (76.8) | 55/69 (79.7) | 18/26 (69.2) | 0.28 |
| Current smoking | 19/95 (20.0) | 12/69 (17.4) | 7/26 (26.9) | 0.30 |
| Cardiovascular history (%) | ||||
| Previous MI | 19/95 (20.0) | 14/69 (20.3) | 5/26 (19.2) | 0.91 |
| Previous PCI | 13/95 (13.7) | 10/69 (14.5) | 3/26 (11.5) | 0.69 |
| Previous CABG | 4/95 (4.2) | 2/69 (2.9) | 2/26 (7.7) | 0.31 |
| Previous medication (%) | ||||
| Aspirin | 38/95 (40.0) | 27/69 (39.1) | 11/26 (42.3) | 0.78 |
| Clopidogrel | 16/95 (16.8) | 5/26 (19.2) | 11/69 (15.9) | 0.70 |
| Beta-blockers | 23/95 (24.2) | 19/69 (27.5) | 4/26 (15.4) | 0.22 |
| ACE inhibitors | 46/95 (48.4) | 33/69 (47.8) | 13/26 (50.0) | 0.85 |
| Statins | 44/95 (46.3) | 35/69 (50.7) | 9/26 (34.6) | 0.16 |
| Diuretics | 16/95 (16.8) | 14/69 (20.3) | 2/26 (7.7) | 0.14 |
| Nitrates | 8/95 (8.4) | 6/69 (8.7) | 2/26 (7.7) | 0.88 |
ACE: angiotensin-converting enzyme; CABG: coronary artery bypass grafting; IQ: interquartile; MI: myocardial infarction; NSTEMI: non-ST elevation myocardial infarction; PCI: percutaneous coronary intervention; STEMI: ST elevation myocardial infarction; UA: unstable angina.
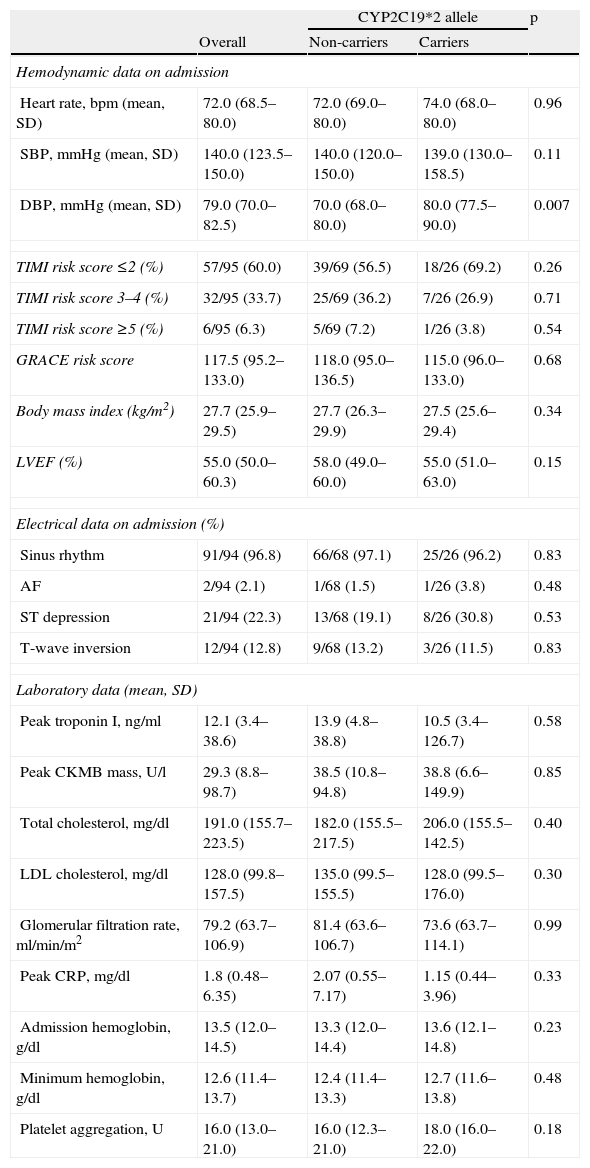
The groups were similar regarding risk assessment according to the TIMI and GRACE risk scores. Other variables such as body weight, left ventricular ejection fraction, peak troponin I, admission and nadir hemoglobin, and glomerular filtration rate were also homogenous for both populations. Platelet function assessed at discharge was similar for both groups (Table 2).
Hemodynamic, electrical and laboratory data.
| CYP2C19*2 allele | p | |||
| Overall | Non-carriers | Carriers | ||
| Hemodynamic data on admission | ||||
| Heart rate, bpm (mean, SD) | 72.0 (68.5–80.0) | 72.0 (69.0–80.0) | 74.0 (68.0–80.0) | 0.96 |
| SBP, mmHg (mean, SD) | 140.0 (123.5–150.0) | 140.0 (120.0–150.0) | 139.0 (130.0–158.5) | 0.11 |
| DBP, mmHg (mean, SD) | 79.0 (70.0–82.5) | 70.0 (68.0–80.0) | 80.0 (77.5–90.0) | 0.007 |
| TIMI risk score ≤2 (%) | 57/95 (60.0) | 39/69 (56.5) | 18/26 (69.2) | 0.26 |
| TIMI risk score 3–4 (%) | 32/95 (33.7) | 25/69 (36.2) | 7/26 (26.9) | 0.71 |
| TIMI risk score ≥5 (%) | 6/95 (6.3) | 5/69 (7.2) | 1/26 (3.8) | 0.54 |
| GRACE risk score | 117.5 (95.2–133.0) | 118.0 (95.0–136.5) | 115.0 (96.0–133.0) | 0.68 |
| Body mass index (kg/m2) | 27.7 (25.9–29.5) | 27.7 (26.3–29.9) | 27.5 (25.6–29.4) | 0.34 |
| LVEF (%) | 55.0 (50.0–60.3) | 58.0 (49.0–60.0) | 55.0 (51.0–63.0) | 0.15 |
| Electrical data on admission (%) | ||||
| Sinus rhythm | 91/94 (96.8) | 66/68 (97.1) | 25/26 (96.2) | 0.83 |
| AF | 2/94 (2.1) | 1/68 (1.5) | 1/26 (3.8) | 0.48 |
| ST depression | 21/94 (22.3) | 13/68 (19.1) | 8/26 (30.8) | 0.53 |
| T-wave inversion | 12/94 (12.8) | 9/68 (13.2) | 3/26 (11.5) | 0.83 |
| Laboratory data (mean, SD) | ||||
| Peak troponin I, ng/ml | 12.1 (3.4–38.6) | 13.9 (4.8–38.8) | 10.5 (3.4–126.7) | 0.58 |
| Peak CKMB mass, U/l | 29.3 (8.8–98.7) | 38.5 (10.8–94.8) | 38.8 (6.6–149.9) | 0.85 |
| Total cholesterol, mg/dl | 191.0 (155.7–223.5) | 182.0 (155.5–217.5) | 206.0 (155.5–142.5) | 0.40 |
| LDL cholesterol, mg/dl | 128.0 (99.8–157.5) | 135.0 (99.5–155.5) | 128.0 (99.5–176.0) | 0.30 |
| Glomerular filtration rate, ml/min/m2 | 79.2 (63.7–106.9) | 81.4 (63.6–106.7) | 73.6 (63.7–114.1) | 0.99 |
| Peak CRP, mg/dl | 1.8 (0.48–6.35) | 2.07 (0.55–7.17) | 1.15 (0.44–3.96) | 0.33 |
| Admission hemoglobin, g/dl | 13.5 (12.0–14.5) | 13.3 (12.0–14.4) | 13.6 (12.1–14.8) | 0.23 |
| Minimum hemoglobin, g/dl | 12.6 (11.4–13.7) | 12.4 (11.4–13.3) | 12.7 (11.6–13.8) | 0.48 |
| Platelet aggregation, U | 16.0 (13.0–21.0) | 16.0 (12.3–21.0) | 18.0 (16.0–22.0) | 0.18 |
AF: atrial fibrillation; CRP: C-reactive protein; DBP: diastolic blood pressure; LVEF: left ventricular ejection fraction; SBP: systolic blood pressure.
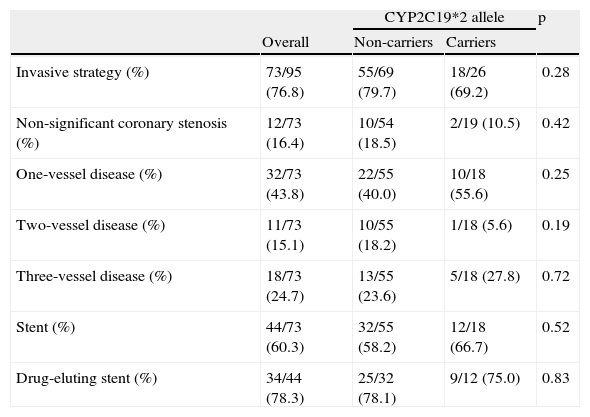
Almost three-quarters of both sub-populations were treated by an invasive strategy. Most had single-vessel disease, and drug-eluting stents were used in both groups (Table 3).
In-hospital management.
| CYP2C19*2 allele | p | |||
| Overall | Non-carriers | Carriers | ||
| Invasive strategy (%) | 73/95 (76.8) | 55/69 (79.7) | 18/26 (69.2) | 0.28 |
| Non-significant coronary stenosis (%) | 12/73 (16.4) | 10/54 (18.5) | 2/19 (10.5) | 0.42 |
| One-vessel disease (%) | 32/73 (43.8) | 22/55 (40.0) | 10/18 (55.6) | 0.25 |
| Two-vessel disease (%) | 11/73 (15.1) | 10/55 (18.2) | 1/18 (5.6) | 0.19 |
| Three-vessel disease (%) | 18/73 (24.7) | 13/55 (23.6) | 5/18 (27.8) | 0.72 |
| Stent (%) | 44/73 (60.3) | 32/55 (58.2) | 12/18 (66.7) | 0.52 |
| Drug-eluting stent (%) | 34/44 (78.3) | 25/32 (78.1) | 9/12 (75.0) | 0.83 |
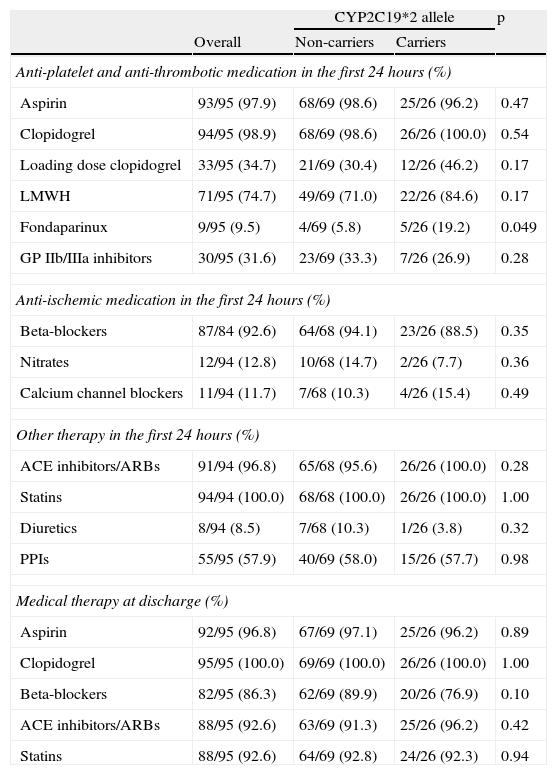
With respect to medical therapy, almost all of the population (97.9%) started on clopidogrel and aspirin during the first 24hours. Only a third of the patients received a loading dose of clopidogrel. The most used anticoagulant was enoxaparin. Fondaparinux was used less frequently in non-carriers (5.8 vs. 19.2%, p=0.049). There were no significant differences regarding medication in the first 24hours or at discharge (Table 4).
Medical therapy.
| CYP2C19*2 allele | p | |||
| Overall | Non-carriers | Carriers | ||
| Anti-platelet and anti-thrombotic medication in the first 24hours (%) | ||||
| Aspirin | 93/95 (97.9) | 68/69 (98.6) | 25/26 (96.2) | 0.47 |
| Clopidogrel | 94/95 (98.9) | 68/69 (98.6) | 26/26 (100.0) | 0.54 |
| Loading dose clopidogrel | 33/95 (34.7) | 21/69 (30.4) | 12/26 (46.2) | 0.17 |
| LMWH | 71/95 (74.7) | 49/69 (71.0) | 22/26 (84.6) | 0.17 |
| Fondaparinux | 9/95 (9.5) | 4/69 (5.8) | 5/26 (19.2) | 0.049 |
| GP IIb/IIIa inhibitors | 30/95 (31.6) | 23/69 (33.3) | 7/26 (26.9) | 0.28 |
| Anti-ischemic medication in the first 24hours (%) | ||||
| Beta-blockers | 87/84 (92.6) | 64/68 (94.1) | 23/26 (88.5) | 0.35 |
| Nitrates | 12/94 (12.8) | 10/68 (14.7) | 2/26 (7.7) | 0.36 |
| Calcium channel blockers | 11/94 (11.7) | 7/68 (10.3) | 4/26 (15.4) | 0.49 |
| Other therapy in the first 24hours (%) | ||||
| ACE inhibitors/ARBs | 91/94 (96.8) | 65/68 (95.6) | 26/26 (100.0) | 0.28 |
| Statins | 94/94 (100.0) | 68/68 (100.0) | 26/26 (100.0) | 1.00 |
| Diuretics | 8/94 (8.5) | 7/68 (10.3) | 1/26 (3.8) | 0.32 |
| PPIs | 55/95 (57.9) | 40/69 (58.0) | 15/26 (57.7) | 0.98 |
| Medical therapy at discharge (%) | ||||
| Aspirin | 92/95 (96.8) | 67/69 (97.1) | 25/26 (96.2) | 0.89 |
| Clopidogrel | 95/95 (100.0) | 69/69 (100.0) | 26/26 (100.0) | 1.00 |
| Beta-blockers | 82/95 (86.3) | 62/69 (89.9) | 20/26 (76.9) | 0.10 |
| ACE inhibitors/ARBs | 88/95 (92.6) | 63/69 (91.3) | 25/26 (96.2) | 0.42 |
| Statins | 88/95 (92.6) | 64/69 (92.8) | 24/26 (92.3) | 0.94 |
ACE: angiotensin-converting enzyme; ARBs: angiotensin receptor blockers; GP: glycoprotein; LMWH: low molecular weight heparin; PPI: proton pump inhibitors.
Follow-up data were available for 91 patients (4.2% lost to follow-up). Median follow-up for the whole population was 136.0 (79.0–188.0) days.
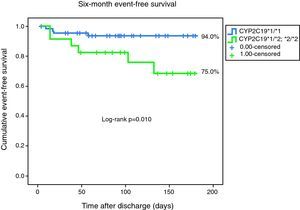
Ten primary endpoint events were recorded during follow-up. One patient died, three had a non-fatal myocardial infarction, and six were readmitted for unstable angina during follow-up. The primary endpoint was reached by four of the 67 homozygous patients, as opposed to six of the 24 heterozygotes. Event-free survival curves for both groups are presented in Fig. 1. This result was significant (log-rank p=0.010).
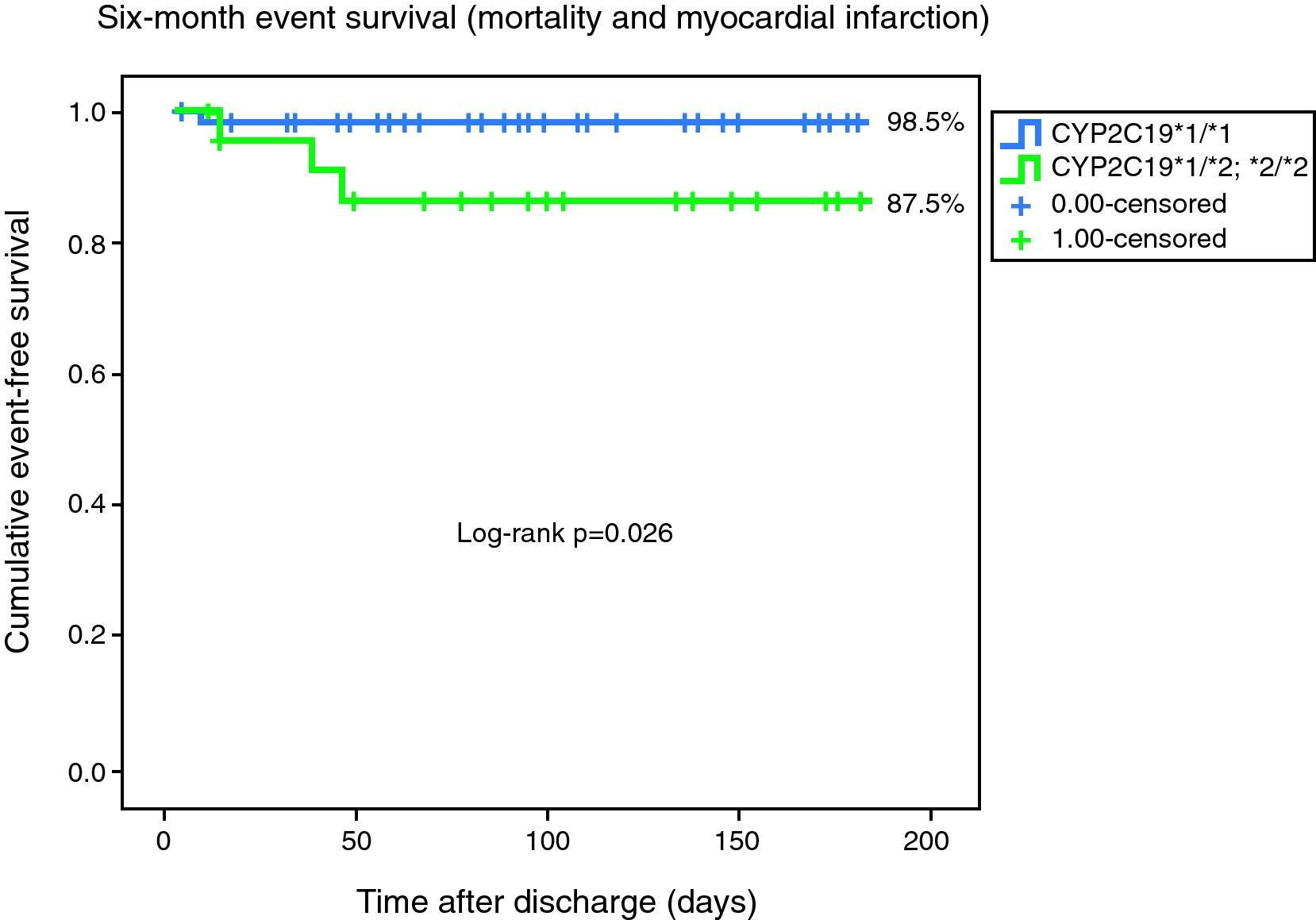
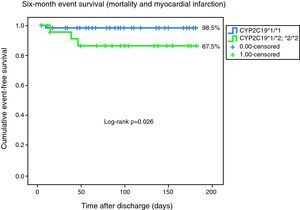
With analysis based solely on the combined outcome of cardiovascular mortality and non-fatal myocardial infarction, freedom from the combined event remained significantly lower for the *2 carrier population (98.5 vs. 87.5%, log-rank p=0.026) (Fig. 2).
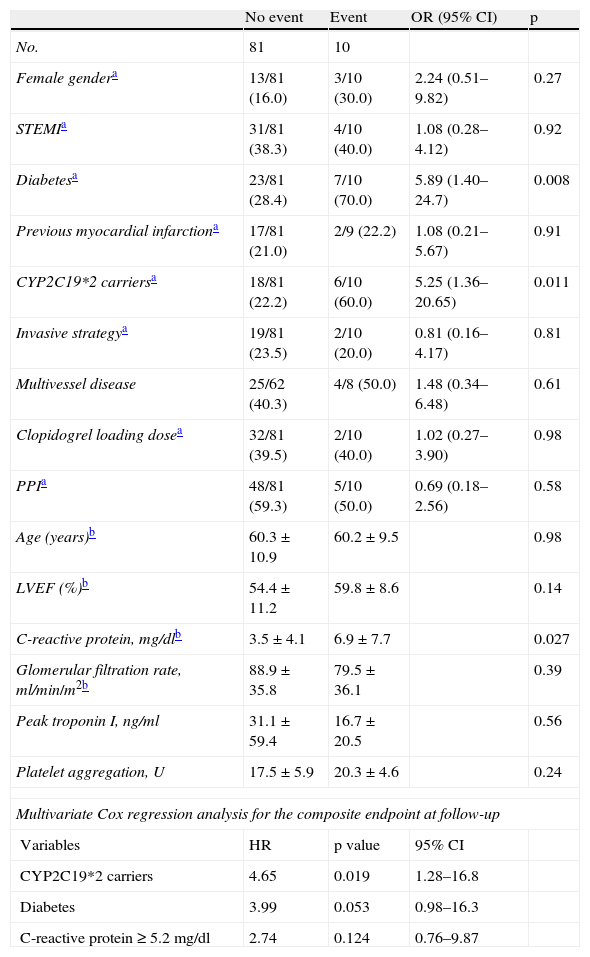
In univariate analysis of the combined endpoint, only the presence of CYP2C19*2, diabetes, and a higher level of C-reactive protein (CRP) were significant. According to the Cox regression model, only the presence of CYP2C19*2 was an independent predictor of outcome, conferring a 4.65 increase in relative risk (95% confidence interval [CI] 1.28–16.8, p=0.019) (Table 5).
Univariate predictors of the combined event.
| No event | Event | OR (95% CI) | p | |
| No. | 81 | 10 | ||
| Female gendera | 13/81 (16.0) | 3/10 (30.0) | 2.24 (0.51–9.82) | 0.27 |
| STEMIa | 31/81 (38.3) | 4/10 (40.0) | 1.08 (0.28–4.12) | 0.92 |
| Diabetesa | 23/81 (28.4) | 7/10 (70.0) | 5.89 (1.40–24.7) | 0.008 |
| Previous myocardial infarctiona | 17/81 (21.0) | 2/9 (22.2) | 1.08 (0.21–5.67) | 0.91 |
| CYP2C19*2 carriersa | 18/81 (22.2) | 6/10 (60.0) | 5.25 (1.36–20.65) | 0.011 |
| Invasive strategya | 19/81 (23.5) | 2/10 (20.0) | 0.81 (0.16–4.17) | 0.81 |
| Multivessel disease | 25/62 (40.3) | 4/8 (50.0) | 1.48 (0.34–6.48) | 0.61 |
| Clopidogrel loading dosea | 32/81 (39.5) | 2/10 (40.0) | 1.02 (0.27–3.90) | 0.98 |
| PPIa | 48/81 (59.3) | 5/10 (50.0) | 0.69 (0.18–2.56) | 0.58 |
| Age (years)b | 60.3±10.9 | 60.2±9.5 | 0.98 | |
| LVEF (%)b | 54.4±11.2 | 59.8±8.6 | 0.14 | |
| C-reactive protein, mg/dlb | 3.5±4.1 | 6.9±7.7 | 0.027 | |
| Glomerular filtration rate, ml/min/m2b | 88.9±35.8 | 79.5±36.1 | 0.39 | |
| Peak troponin I, ng/ml | 31.1±59.4 | 16.7±20.5 | 0.56 | |
| Platelet aggregation, U | 17.5±5.9 | 20.3±4.6 | 0.24 | |
| Multivariate Cox regression analysis for the composite endpoint at follow-up | ||||
| Variables | HR | p value | 95% CI | |
| CYP2C19*2 carriers | 4.65 | 0.019 | 1.28–16.8 | |
| Diabetes | 3.99 | 0.053 | 0.98–16.3 | |
| C-reactive protein≥5.2mg/dl | 2.74 | 0.124 | 0.76–9.87 | |
Chi-square 16.9; p<0.01.
LVEF: left ventricular ejection fraction; PPI: proton pump inhibitor; STEMI: ST elevation myocardial infarction.
Our study confirms the prognostic importance of the *2 polymorphism in CYP2C19, as in a Portuguese single-center unselected ACS population discharged on double antiplatelet therapy, this SNP conferred less protection from recurrent cardiac ischemic events.
CYP2C19*2 frequencyOur local data were in agreement with previous studies on Caucasians populations that reported *2 frequencies ranging from 11.1 to 15.9%.16–18 Our CYP2C19*2 frequency of 14.2% was lower than others, particularly in an Asian cohort in which the frequency of CYP2C19*2 reached 45%.19 The CYP2C19*3 allele was not found in any patient in our cohort (data not shown), which has also been reported in Caucasian cohorts by Scordo et al.16 and Goldstein et al.18
MechanismThere are compelling biologic data to support the association between CYP2C19*2 and prognosis. CYP2C19 contributes to both of the sequential oxidative metabolic steps of clopidogrel activation. Slowing the first reaction would tend to shunt the prodrug preferentially to an esterase-mediated pathway forming pharmacologically inactive metabolites.20 Although there are many genetic polymorphisms of CYP2C19, the *2 allele was the most frequent variant in the reduced-function group. This loss-of-function variant encodes a cryptic splice variant that leads to no enzymatic activity.21
Decreased CYP2C19 activity leads to decreased exposure to the active metabolite of clopidogrel and to reduced pharmacodynamic response that is probably the basis of HPR.15,22 HPR has been documented after a loading dose of clopidogrel, but also after 75mg clopidogrel per day for one week.23 Our sample size was probably the main reason for the fact that ADP-induced platelet aggregation was not statistically different in the two patient groups.
Another explanation for the prognostic impact of the SNP studied was the association between CYP2C19*2 and higher concentrations of inflammatory markers, notably interleukin-6 and high sensitivity CRP.24 CYP2C19*2 could therefore affect underlying cardiovascular risk in addition to its pharmacogenetic effect on clopidogrel.25 However, in the FAST-MI trial, no such effect was seen in the subgroup of 222 patients who contributed a blood sample to the DNA bank but who did not receive clopidogrel,4 suggesting that the prognostic implications of the CYP2C19 polymorphism were probably due to its influence on clopidogrel metabolism.
Prognostic implicationsThe magnitude of the effect of carriage of the CYP2C19*2 allele on prognosis in our population was higher than expected from recent trials20 and even from a collaborative metanalysis26 (HR 1.6 for an endpoint of major cardiovascular events with respect to carriers of the *2 allele vs. non-carriers). Nevertheless, our data were similar to those of Collet et al.,25 in which carriers of the CYP2C19*2 allele had a four times higher rate of the cardiac ischemic endpoint (HR 4.6 for an endpoint of cardiovascular death, myocardial infarction, and myocardial revascularization).
By contrast, previous trials concluded that CYP2C19*2 by itself was not an independent predictor of outcome, among them the FAST-MI study, in which no SNPs in CYP2C19, or in CYP3A5, P2RY12, or ITGB3, were associated with the primary outcome. Of note and probably explaining these results, many demographic and clinical characteristics differed between patients who had the primary outcome and those who did not. Specifically, those that had the primary outcome (294 patients [13%]) were older, had more comorbidities, were more likely to be receiving drugs for secondary prevention (including clopidogrel) before their infarction, and were less likely to receive primary PCI or intravenous fibrinolysis during the inclusion event.4
Reports from substudies of the EXCELSIOR (Impact of Extent of Clopidogrel-Induced Platelet Inhibition During Elective Stent Implantation on Clinical Event Rate) and CLARITY-TIMI 28 (Clopidogrel as Adjunctive Reperfusion Therapy-Thrombolysis in Myocardial Infarction) trials also found no association between CYP2C19 genotype and prognosis.27,28
It also seems that, as shown by the collaborative meta-analysis, CYP2C19*2 has a greater impact in the context of an ACS and percutaneous revascularization, in terms not only of stent thrombosis but also of cardiac ischemic endpoints.26 In FAST-MI and CLARITY-TIMI 28, trials in which carriers of the CYP2C19*2 did not have a worse prognosis, only 70% and 58% of patients, respectively, underwent PCI.4,27
Future prospectsThere appears to be agreement that CYP2C19*2 carriers have worse ischemic outcomes and are more prone to stent thrombosis.
One question that arises is who should be tested. There is no clear answer to this in the literature, but patients with a history of stent thrombosis, premature cardiovascular disease, or frequent readmissions for ACS would appear to be candidates for genetic testing.
Another issue is how to manage CYP2C19*2 and clopidogrel therapy. Should the daily dosage be increased? Should a different antiplatelet agent such as prasugrel be used? At present there is insufficient evidence to answer to this question.
Finally, the interplay between platelet function testing and clopidogrel genotyping is still not clear. Should platelet function testing be used to determine whether genome studies are required? Or, on the contrary, should platelet function be assessed only after genotyping indicates the need?
In the future, large-scale trials to test genetic strategies could provide answers to these questions.
LimitationsPlasma concentrations of the active metabolite of clopidogrel (R130,964) were not measured. The small population size made it impossible to determine the prognostic importance of CYP2C19 genotyping regarding other outcomes such as stent thrombosis and bleeding complications. Sample size may also have been responsible for the fact that platelet function tests did not differ between the groups. The low event rate negatively influenced the quality of the multivariate analysis. Drug compliance during follow-up was not assessed.
ConclusionsIn a single-center ACS population, the CYP2C19*2 allele was an independent prognostic marker, conferring a 4.7 higher relative risk of a cardiac ischemic endpoint in medium-term follow-up.
Conflicts of interestThe authors have no conflicts of interest to declare.