Dilated cardiomyopathy (DCM) is a disease of the heart muscle characterized by ventricular dilatation and impaired systolic function. Familial forms account for 30-50% of cases. Autosomal dominant inheritance is the predominant pattern of transmission. Causal genetic variants have been identified in several genes and molecular diagnosis has implications for genetic counseling and risk stratification.
ObjectiveWe aimed to estimate the frequency of genetic variants and the molecular basis of DCM in Portugal.
MethodsWe performed a multicenter study of unrelated patients, recruited between 2013 and 2014. Variants in 15 genes were screened using PCR with direct sequencing (next-generation sequencing with at least 30-fold coverage combined with Sanger sequencing).
ResultsA total of 107 patients were included, 64 (60%) men, mean age at diagnosis 38±13 years, with 48 (45%) familial cases. In total, 31 rare variants in eight genes (mainly in MYBPC3, TNNT2 and LMNA) were identified, in 28 patients (26%). Only four variants had been previously described in association with DCM, 11 with hypertrophic cardiomyopathy, and nine variants were novel. Four variants were likely pathogenic and the remainder were of uncertain significance. We found no major differences in the main clinical and imaging characteristics between patients with or without rare variants and patients with likely pathogenic variants.
ConclusionsOur results reflect the complexity and diversity of DCM genetics. For better interpretation of the pathogenicity of the variants found and their causative roles in DCM, molecular cascade screening of families is imperative. Further insight into genotype-phenotype correlations and risk stratification is desirable.
A miocardiopatia dilatada (MCD) é uma doença do músculo cardíaco caracterizada por dilatação ventricular e compromisso da função sistólica. As formas familiares são responsáveis por 30 a 50% dos casos. O padrão de hereditariedade predominante é o autossómico dominante. Variantes genéticas causais foram identificadas em vários genes e o diagnóstico molecular tem implicações para o aconselhamento genético e estratificação de risco.
ObjetivoAvaliar a base molecular da MCD em Portugal.
MétodosEstudo multicêntrico de doentes não relacionados, recrutados entre 2013 e 2014. Foram analisados 15 genes, através da técnica de PCR com sequenciação direta (NGS com pelo menos uma cobertura de 30 vezes combinada com sequenciação de Sanger).
ResultadosForam incluídos 107 pacientes, 64 (60%) homens, idade média ao diagnóstico de 38 ± 13 anos, com 48 (45%) casos familiares. Foram identificadas 31 variantes raras, em oito genes, (principalmente MYBPC3, TNNT2 e LMNA) em 28 pacientes (26%). Apenas quatro variantes tinham sido previamente descritas em associação com MCD, 11 com miocardiopatia hipertrófica e nove variantes eram novas. Quatro variantes foram classificadas como provavelmente patogénicas e as restantes de significado incerto. Não encontrámos diferenças significativas nas principais características clínicas e imagiológicas entre doentes com ou sem variantes raras e doentes com variantes provavelmente patogénicas.
ConclusõesEstes resultados refletem a complexidade e diversidade genética da MCD. Para uma melhor interpretação da patogenicidade das variantes e potencial causalidade, o rastreio molecular das famílias é imperativo. Uma visão mais aprofundada das correlações genótipo-fenótipo e da estratificação de risco é desejável.
Dilated cardiomyopathy (DCM) is a disease of the heart muscle characterized by ventricular dilatation and impaired systolic function, in the absence of abnormal loading conditions or coronary artery disease sufficient to cause global systolic impairment.1 Clinically, DCM is manifested by signs or symptoms of heart failure (HF), ventricular and supraventricular arrhythmias, conduction system abnormalities, thromboembolism or sudden death.2 DCM is a leading cause of HF, with 30-50% of enrolled patients in many HF registries and clinical trials categorized as having non-ischemic DCM.3 DCM is the third most common cause of HF and the main reason for heart transplantation.2
The true prevalence of DCM is difficult to ascertain. Earlier estimates derived from a study performed in 1975-1984 in Olmstead County, MN, USA, indicated a prevalence of approximately 1:2700 individuals,4 which is probably underestimated, due to the less sensitive imaging techniques used at that time.5 More recently, Hershberger et al.,6 using different methods, considered the prevalence to be between 1:250 and 1:400.
Familial forms are assumed to account for 30-50% of cases, with autosomal dominant inheritance being the predominant pattern of transmission, although autosomal recessive, X-linked and mitochondrial inheritance have also been described.5,7–10 In recent years, enormous advances have been made in our knowledge of the genetic basis of DCM, especially with the advent of new sequencing technologies, particularly next-generation sequencing (NGS), which enables several genes to be sequenced simultaneously, with decreased costs and turnaround times. However, accurate molecular diagnosis and determination of genotype-phenotype correlations are hampered by genetic heterogeneity and difficulty in correctly interpreting the clinical impact of variants.
Rare genetic variants in more than 50 disease-related genes coding for proteins of the different cell components (cytoskeleton, sarcomere, nuclear membrane and ion channels) have been identified, with considerable overlap with other cardiomyopathies.6,11–14 Besides this locus heterogeneity, there is also marked allelic heterogeneity, with disease-related variants scattered throughout the causal genes.6,11 Most variants are missense, characterized by variable expressivity6,15 and low and age- and gene-dependent penetrance.6,9,15,16 The majority of disease-causing variants are private (unique to a specific family)7,8,15 and it has been suggested that 9-13% of patients carry at least two potentially disease-causing variants.10,16,17 Nevertheless, genetic diagnosis in DCM is recommended in the international guidelines, since it appears to have implications for clinical practice in genetic counseling and risk stratification.15,18
In this context, we aimed to investigate the genetic background and molecular basis of familial and idiopathic cases of DCM in Portugal and to assess possible genotype-phenotype correlations.
MethodsPatients and clinical assessmentThis was a multicenter study of unrelated adult Portuguese DCM patients, recruited between 2013 and 2014 from 10 hospitals with specialist clinics for the treatment of cardiomyopathies (Supplementary Material, Appendix A). The investigation was conducted in accordance with the principles of the 1975 Declaration of Helsinki, the study was approved by the local ethics committees and all subjects gave written informed consent.
DCM was diagnosed according to the criteria of the European Society of Cardiology's Working Group on Myocardial and Pericardial Diseases.1 Specific echocardiographic criteria were the presence of left ventricular systolic dysfunction (ejection fraction <45% and/or fractional shortening <25%) and left ventricular dilatation (end-diastolic diameter ≥98th percentile according to gender and height, from the Framingham Heart Study19).
Familial DCM was diagnosed when there was at least one additional affected family member or if unexplained sudden cardiac death occurred under 35 years of age in a first-degree relative.20 General clinical assessment and cardiological investigations including electrocardiogram and echocardiogram were performed as previously described.21
Molecular diagnosisFor each index case 10 ml of blood was collected in an EDTA tube. Genomic DNA was extracted from peripheral leukocytes in the MagNA Pure device (Roche, Germany), with the MagNA Pure LC DNA Large Volume Isolation Kit (Roche, Germany), according to the manufacturer's instructions. DNA quantity and quality were assessed on a NanoDrop 2000c. The samples were sent by surface mail to a central laboratory (Institute of Molecular Pathology and Immunology at the University of Porto, Portugal), where DNA was extracted for molecular analysis and stored at -70°C.
Variants in a panel of 15 genes (Table 1) were screened in all patients, using NGS with at least 30-fold coverage. Sanger sequencing was used to validate the identified variants and to provide additional coverage for regions of the panel with less than 30-fold coverage. Genes were selected and the targeted gene panel was designed in 2010, based on previously identified variants in DCM patients at the time.
Genes included in the molecular analysis.
| Gene | Protein | Chromosome | OMIM | NCBI reference sequences | |
|---|---|---|---|---|---|
| NM | NP | ||||
| Sarcomere | |||||
| ACTC1 | α-cardiac actin | 15q14 | 102540 | NM_005159.4 | NP_005150.1 |
| MYBPC3 | Cardiac myosin-binding protein C | 11p11.2 | 600958 | NM_000256.3 | NP_000247.2 |
| MYL2 | Myosin light chain 2, regulatory | 12q24.11 | 160781 | NM_000432.3 | NP_000423.2 |
| MYL3 | Myosin light chain 3, essential | 3p21.31 | 160790 | NM_000258.2 | NP_000249.1 |
| MYH7 | β-myosin heavy chain | 14q11.2 | 160760 | NM_000257.3 | NP_000248.2 |
| TNNI3 | Cardiac troponin I | 19q13.42 | 191044 | NM_000363.4 | NP_000354.4 |
| TNNT2 | Cardiac troponin T | 1q32.1 | 191045 | NM_001001430.2 | NP_001001430.1 |
| TPM1 | α-tropomyosin 1 | 15q22.2 | 191010 | NM_000366.5 | NP_000357.3 |
| Nuclear lamina | |||||
| LMNA | Lamin A/C | 1q22 | 150330 | NM_170707.3 | NP_733821.1 |
| Z-disk | |||||
| CSRP3 | Cystein- and glycine-rich protein 3 | 11p15.1 | 600824 | NM_003476.4 | NP_003467.1 |
| LDB3 | LIM domain-binding | 10q23.2 | 605906 | NM_007078.2 | NP_009009.1 |
| TCAP | Titin cap-telethonin | 17q12 | 604488 | NM_003673.3 | NP_003664.1 |
| Dystrophin complex | |||||
| SGCD | δ-sarcoglycan | 5q33.2-q33.3 | 601411 | NM_001128209.1 | NP_001121681.1 |
| Sarcoplasmic reticulum (calcium handling) | |||||
| PLN | Phospholamban | 6q22.31 | 172405 | NM_002667.3 | NP_002658.1 |
| Mitochondria | |||||
| TAZ | Tafazzin | Xq28 | 300394 | NM_000116.4 | NP_000107.1 |
NCBI: National Center for Biotechnology Information; NM: mRNA reference sequence; NP: protein reference sequence; OMIM: Online Mendelian Inheritance in Man.
Reads were assembled with SeqMan NGen version 4.1 (DNASTAR, Madison, WI) using the FASTQ files containing sequence reads and the template references (from the hg19 human reference genome) adjusted for the covered amplicons. SeqMan Pro version 10 (DNASTAR) was used as a post-assembly tool for the analysis of overall amplicon coverage, depth of coverage of individual bases, and variant identification. A filter for the coding sequence variants was applied to the SNP report for each case and each variant was interpreted on this basis.
Variant classificationPathogenicity was assessed by comparisons with populational (mainly Exome Aggregation Consortium [ExAC] and Single Nucleotide Polymorphism) and disease databases (the Human Gene Mutation Database [HGMD], ClinVar and the Leiden Open Variation Database) in which genetic variants have been described, predictive bioinformatics programs (PolyPhen-2, SIFT, Mutation Taster and Human Splicing Finder), familial segregation analysis and functional studies when available, in accordance with the American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG/AMP) guidelines.22 Variants were classified as ‘pathogenic’, ‘likely pathogenic’, ‘of uncertain significance’, ‘likely benign’ and ‘benign’.
Family assessmentClinical assessment, electrocardiogram and echocardiogram were performed in all available family members. Subsequently, if a rare genetic variant was found in an index case, the identified variant was screened among all available relatives.
Statistical analysisIBM SPSS 24.0 (IBM SPSS Statistics, 2016) was used for the statistical analysis. Baseline patient characteristics are expressed as mean ± standard deviation or median (interquartile range), and categorical variables as absolute number (percentage). Between-group comparisons were performed using Pearson's chi-square for categorical variables, if appropriate, otherwise Fisher's exact test was used. For continuous variables, the Student's t test or the Mann-Whitney U test were used, according to data distribution. Differences were considered significant for a two-sided p value of <0.05.
ResultsStudy population and clinical characteristicsA total of 107 unrelated DCM patients were included, 64 (60%) men. The mean age at diagnosis was 38±13 years. Familial disease was present in 48 (45%) probands and 25 (24%) had a family history of sudden cardiac death.
The most frequent clinical presentation was HF symptoms (dyspnea, orthopnea or fatigue), present in 73 (70%) patients. Mean left ventricular end-diastolic dimension by echocardiography was 64±9 mm and mean ejection fraction was 31±11%; 25 (26%) patients also presented right ventricular involvement. Sixty-eight (64%) were studied by cardiac magnetic resonance, presenting a mean left ventricular end-diastolic volume index of 132±36ml/m2, and mean ejection fraction was 32±11%; 26 (39%) patients presented some degree of late gadolinium enhancement (LGE), mostly with intramyocardial distribution.
By the time of recruitment, 58 (57%) patients had had at least one cardiac-related hospitalization, 27 (27%) patients had received an implantable cardioverter-defibrillator, 11 (11%) had received cardiac resynchronization therapy, six (6%) a conventional pacemaker and 11 (10%) had undergone heart transplantation. Table 2 details the main clinical and imaging characteristics of the study population.
Characteristics of patients with dilated cardiomyopathy (n=107).
| Gender, n (%) | |
| Male | 64 (60) |
| Female | 43 (40) |
| Age at diagnosis, years (mean ± SD) | 38±13 |
| Familial DCM, n (%) | 48 (45) |
| Idiopathic DCM, n (%) | 59 (55) |
| NYHA functional class, n (%) | |
| I | 47 (49) |
| II | 44 (45) |
| III | 5 (5) |
| IV | 1 (1) |
| ECG data, n (%) | |
| LBBB | 40 (38) |
| AF/atrial flutter | 11 (11) |
| NSVT | 19 (26) |
| Echocardiographic data | |
| LVEDD, mm (mean ± SD) | 64±9 |
| LVEF, % (mean ± SD) | 31±11 |
| RV impairment, n (%) | 25 (26) |
| CMR data | |
| LVEDV, ml/m2 (mean ± SD) | 132±36 |
| LVEF, % (mean ± SD) | 32±11 |
| LGE, n (%) | 26 (39) |
| ICD implantation, n (%) | 27 (27) |
| CRT implantation, n (%) | 11 (11) |
| Heart transplantation, n (%) | 11 (10) |
AF: atrial fibrillation; CMR: cardiac magnetic resonance; CRT: cardiac resynchronization therapy; DCM: dilated cardiomyopathy; ECG: electrocardiogram; ICD: implantable cardioverter-defibrillator; LBBB: left bundle branch block; LGE: late gadolinium enhancement; LVEDD: left ventricular end-diastolic diameter; LVEDV: left ventricular end-diastolic volume; LVEF: left ventricular ejection fraction; NSVT: non-sustained ventricular tachycardia; NYHA: New York Heart Association; RV: right ventricle; SD: standard deviation.
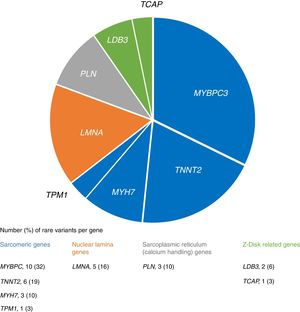
We identified 31 rare variants in eight genes (mainly in MYBPC3, TNNT2 and LMNA) in 28 (26%) patients, most frequently in sarcomeric genes (Figure 1). In familial cases there was a higher proportion of patients with rare variants (33% vs. 20%), although without statistical significance (p=0.128).
Approximately one third of the variants (9/31) had never before been described (novel variants). Eleven had been previously described in association with hypertrophic cardiomyopathy (HCM) (mainly in MYBPC3), four in association with DCM (in LMNA and TNNT2), four with left ventricular non-compaction and one with restrictive cardiomyopathy (Table 3). Five patients had more than one rare variant (including cases of compound heterozygosity and double heterozygosity) and two rare variants were present in more than one proband (Table 4 and Figure 2A).
Rare genetic variants (compared to hg19 reference genome) and previously associated phenotype.
| Gene | Variant | dbSNP | Associated phenotype |
|---|---|---|---|
| MYBPC3 | c.1226+6 T>C | rs397515892 | HCM (ClinVar) |
| c.50 G>A, p.Arg17Gln | rs374630007 | HCM23 | |
| c.131 G>A, p.Arg44His | rs369205562 | – | |
| c.223 G>A, p.Asp75Asn | rs375471260 | HCM (ClinVar) | |
| c.833 G>A, p.Gly278Glu | rs147315081 | HCM (ClinVar) | |
| c.836 G>C, p.Gly279Ala | rs375774648 | HCM (ClinVar) | |
| c.1298 C>G, p.Ala433Gly* | – | ||
| c.1321 G>A, p.Glu441Lys | rs193922377 | HCM (ClinVar) Wolff-Parkinson-White (ClinVar) | |
| c.1484 G>A, p.Arg495Gln | rs200411226 | HCM(ClinVar) | |
| c.1855 G>A, p.Glu619Lys | rs200352299 | HCM (ClinVar) LVNC (ClinVar) | |
| TNNT2 | c.83 C>T, p.Ala28Val | rs200754249 | Increased left ventricular thickness (ClinVar) Familial RCM (ClinVar) HCM (ClinVar) LVNC (ClinVar) |
| c.325 C>T, p.His109Tyra | rs397516460 | – | |
| c.517 C>T, p.Arg173Trp | rs727503512 | DCM (ClinVar) LVNC (ClinVar) | |
| c.640 G>A, p.Glu214Lys | rs769128006 | – | |
| c.808 G>C, p.Asp270His* | – | ||
| c.824 C>T, p.Ser275Phe* | – | ||
| LMNA | c.460 G>A, p.Glu154Lys* | – | |
| c.1003 C>T, p.Arg335Trp | rs386134243 | DCM (ClinVar) Laminopathies (ClinVar) Hutchinson-Gilford syndrome (ClinVar) Familial partial lipodystrophy (ClinVar) | |
| c.1318 G>A, p.Val440Met | rs121912493 | Partial lipodystrophy (ClinVar) Laminopathies (ClinVar) | |
| c.1604 G>C, p.Gly535Ala | rs747717293 | – | |
| c.1982 G>C, p.Cys661Ser* | – | ||
| MYH7 | c.3942 C>G, p.Asp1314Glu* | – | |
| c.4377 G>T, p.Lys1459Asn | rs201307101 | HCM (ClinVar) Wolff-Parkinson-White (ClinVar) | |
| c.5623 G>T, p.Val1875Phe* | – | ||
| PLN | c.23 C>T, p.Thr8Ile* | – | |
| c.26 G>A, p.Arg9His | rs754782171 | DCM24 | |
| c.61 C>A, p.Pro21Thr | rs397516786 | – | |
| LDB3 | c.466 G>A, p.Ala156Thr | rs200596619 | LVNC (ClinVar) Myofibrillar myopathy (ClinVar) |
| c.1189 G>A, p.Val397Ile | rs773827529 | – | |
| TCAP | c.313 G>C, p.Glu105Glnb | rs146906267 | DCM (ClinVar) HCM (ClinVar) |
| TPM1 | c.758 T>C, p.Ile253Thr* | – |
DCM: dilated cardiomyopathy; dbSNP: Single Nucleotide Polymorphism Database; HCM: hypertrophic cardiomyopathy; LVNC: left ventricular noncompaction; RCM: restrictive cardiomyopathy.
Variants in bold: likely pathogenic variants.
Multiple rare variants affecting single patients.
| No. of variants per patient | Variants |
|---|---|
| 2 | MYBPC3 (c.836G>C, p.Gly279Ala) |
| MYBPC3 (c.1321G>A, p.Glu441Lys)a | |
| LDB3 (c.466G>A, p.Ala156Thr) | |
| LDB3 (c.1189G>A, p.Val397Ile) | |
| TNNT2 (c.325C>T, p.His109Tyr) | |
| TCAP (c.313G>C, p.Glu105Gln) | |
| MYBPC3 (c.131G>A, p.Arg44His) | |
| TNNT2 (c.824C>T, p.Ser275Phe) | |
| 3 | LMNA (c.1318G>A, p.Val440Met) |
| LMNA (c.1604G>C, p.Gly535Ala) | |
| LMNA (c.1982G>C, p.Cys661Ser) |
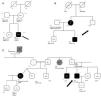
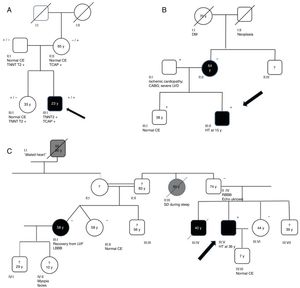
Genograms in three families of dilated cardiomyopathy patients. (A) Family with multiple variants in TNNT2 (c.325C>T, p.His109Tyr) and TCAP (c.313G>C, p.Glu105Gln), in which the phenotype is only expressed in cases of double heterozygosity, and the presence of just one variant is not associated with the expression of DCM; (B and C) families with non-segregating TNNT2 variant (c.325C>T, p.His109Tyr) and LMNA variant (c.460G>A, p.Glu154Lys), respectively. Square: male; circle: female; black symbol: dilated cardiomyopathy; gray symbol: probable cardiac pathology; crossed symbol: deceased family member; arrow: proband; +/− symbols: presence/absence of the variant; numbers inside the symbols: age (years); ?: unavailability for clinical/genetic assessment; CABG: coronary artery bypass grafting; CE: cardiac evaluation; HT: heart transplantation; LBBB: left bundle branch block; LVD: left ventricular dysfunction; LVF: left ventricular function; RBBB: right bundle branch block; SD: sudden death; y: years.
Family segregation analysis was performed whenever possible, but was restricted by the small size of the families and lack of data. Consequently, it was only possible to demonstrate the absence of segregation in three families, with variants in the TNNT2, LMNA (Figure 2B and C) and TCAP25 genes.
Applying the ACMG/AMP guidelines,22 four variants (13%) were classified as likely pathogenic and the other 27 (87%) as of uncertain significance (Table 5).
Likely pathogenic variants according to American College of Medical Genetics and Genomics/Association for Molecular Pathology criteria.
| Gene | Transcript | Nucleotide change | Amino acid change | ACMG/AMP criteriaa | Type of DCM |
|---|---|---|---|---|---|
| MYBPC3 | ENST00000545968 | c.1321G>A | p.Glu441Lys | PS4, PP3, PP5 | Familial |
| MYBPC3 | ENST00000545968 | c.1484G>A | p.Arg495Gln | PS4, PP3, PP1 | Familial |
| TNNT2 | ENST00000367318 | c.517C>T | p.Arg173Trp | PS4, PP3, PP1, PP5, PP4 | Idiopathic |
| LMNA | ENSG00000160789 | c.1003C>T | p.Arg335Trp | PS4, PP3, PP5, PP4 | Idiopathic |
ACMG/AMP: American College of Medical Genetics and Genomics/Association for Molecular Pathology; DCM: dilated cardiomyopathy.
Evidence for classification of variants: PS: pathogenic strong; PP: pathogenic supporting; the numbering within each category does not convey any differences of weight and refer the different criteria: PS4: the prevalence of the variant in affected individuals is significantly increased compared with the prevalence in controls; PP1: co-segregation with disease in multiple affected family members in a gene definitively known to cause the disease; PP3: multiple lines of computational evidence support a deleterious effect on the gene or gene product; PP4: patient's phenotype or family history is highly specific for a disease with a single genetic etiology; PP5: reputable source recently reports variant as pathogenic but the evidence is not available to the laboratory to perform an independent evaluation.22
The main clinical and imaging characteristics of patients with a rare variant were not statistically different from those without a rare variant (Table 6). Female patients tended to present more frequently with left bundle branch block (52% vs. 29%, p=0.016), to have a higher body mass index (28±6 kg/m2 vs. 26±3 kg/m2, p=0.033) and to be more symptomatic (NYHA class >I in 65% vs. 42%, p=0.026), although male patients had more previous hospitalizations (67% vs. 42%, p=0.010).
Patient characteristics according to molecular study results.
| Patient characteristic | No variant (n=79) | Variant-positive (n=28) | p | Likely pathogenic variant (n=4) | p |
|---|---|---|---|---|---|
| Age, years (mean ± SD) | 46±11 | 49±11 | 0.240 | 48±10 | 0.759 |
| Age at diagnosis, years (mean ± SD) | 38±14 | 39±12 | 0.871 | 40±7 | 0.824 |
| Male, n (%) | 48 (61) | 16 (57) | 0.737 | 4 (100) | 0.292 |
| Familial cases, n (%) | 32 (40) | 16 (57) | 0.128 | 2 (50) | 1.000 |
| Family history, n (%) | |||||
| HF-related death | 21 (28) | 5 (19) | 0.396 | 0 (0) | 0.568 |
| Heart transplant | 2 (3) | 1 (4) | 1.000 | 0 (0) | 1.000 |
| Sudden cardiac death | 19 (24) | 6 (23) | 0.919 | 0 (0) | 0.569 |
| Clinical presentation, n (%) | |||||
| HF | 55 (71) | 18 (67) | 0.491 | 4 (100) | 0.298 |
| Syncope/arrhythmia | 14 (18) | 1(4) | 0.051 | 0 (0) | 0.668 |
| NYHA >I, n (%) | 36 (50) | 14 (56) | 0.605 | 3 (75) | 0.615 |
| Previous hospitalizations, n (%) | 44 (57) | 14 (56) | 0.920 | 3 (75) | 0.635 |
| HF | 33 (43) | 7 (28) | 0.186 | 0 (0) | 0.142 |
| Arrhythmic | 13 (17) | 7 (28) | 0.252 | 3 (75) | 0.023 |
| Heart transplant, n (%) | 7 (9) | 4 (15) | 0.468 | 0 (0) | 1.000 |
| Age, years (mean ± SD) | 37±13 | 38±17 | 0.889 | – | – |
| ECG data, n (%) | |||||
| LBBB | 27 (35) | 13 (50) | 0.163 | 2 (50) | 0.612 |
| Any conduction disordera | 42 (54) | 14 (54) | 1.000 | 3 (75) | 0.623 |
| AF/atrial flutter | 8 (10) | 3 (12) | 1.000 | 1 (25) | 0.378 |
| Any SVTb | 19 (24) | 8 (30) | 0.566 | 2 (50) | 0.264 |
| NSVT | 15 (28) | 4 (21) | 0.538 | 1 (33) | 1.000 |
| Echocardiographic data | |||||
| LVEDD, mm (mean ± SD) | 64±9 | 63±8 | 0.618 | 59±7 | 0.264 |
| LVEF, % (mean ± SD) | 30±11 | 34±11 | 0.116 | 36±7 | 0.284 |
| RV impairment, n (%) | 16 (22) | 9 (36) | 0.187 | 3 (75) | 0.048 |
| CMR data | |||||
| LVEDV, ml/m2 (mean ± SD) | 130±36 | 137±36 | 0.481 | 113±6 | 0.519 |
| LVEF, % (mean ± SD) | 32±11 | 33±11 | 0.785 | 32±5 | 0.947 |
| LGE, n (%) | 19 (37) | 7 (44) | 0.642 | 2 (100) | 0.152 |
| Devices, n (%) | |||||
| ICD | 18 (24) | 9 (36) | 0.242 | 2 (50) | 0.264 |
| CRT | 9 (12) | 2 (8) | 0.726 | 0 (0) | 1.000 |
| PM | 5 (7) | 1 (4) | 1.000 | 0 (0) | 1.000 |
AF: atrial fibrillation; CMR: cardiac magnetic resonance; CRT: cardiac resynchronization therapy; ECG: electrocardiogram; HF: heart failure; ICD: implantable cardioverter-defibrillator; LBBB: left bundle branch bock; LGE: late gadolinium enhancement; LVEDD: left ventricular end-diastolic diameter; LVEDV: left ventricular end-diastolic volume; LVEF: left ventricular ejection fraction; NSVT: non-sustained ventricular tachycardia; NYHA: New York Heart Association functional class; PM: pacemaker; RV: right ventricle; SD: standard deviation; SVT: supraventricular tachycardia.
We compared patients with variants classified as likely pathogenic with those without rare variants (Table 6). Patients with these variants had a higher proportion of previous hospitalizations for arrhythmic causes (75% vs. 17%, p=0.023) and right ventricular impairment on echocardiography (75% vs. 22%, p=0.048); there were no statistically significant differences in terms of the other main clinical and imaging characteristics. Additionally, we analyzed patients with MYBPC3 (n=9), TNNT2 (n=7), MYH7 (n=3), LMNA (n=3) and PLN (n=3) variants and compared them to those with a negative molecular study. There were no statistically significant differences in demographic, clinical or familial characteristics, implantation of devices or ECG abnormalities, although TNNT2 and MYH7 variant carriers had a younger and older age of presentation, respectively, relative to other genetic variant carriers (31±17 years and 48±7 years, respectively). Patients with TNNT2 and MYH7 variants presented significantly higher left ventricular ejection fraction (40±9% and 43±9% vs. 30±11%, p=0.016 and p=0.043, respectively) and patients with LMNA variants presented a higher proportion of right ventricular impairment (100% vs. 22%, p=0.015), compared with patients without a rare variant.
In our cohort, we found multiple rare variants affecting five distinct patients (Table 4), all male. In two cases (one patient with three LMNA variants and one with double LDB3 variants), the clinical scenario was particularly dismal, with previous multiple hospitalizations, implantable cardioverter-defibrillator implantation and heart transplantation.
DiscussionIn this study we investigated the genetic background and molecular basis of familial and idiopathic cases of DCM in Portugal, using NGS, which enabled us to study a relatively large panel of genes, more efficiently and at lower cost, compared to previous sequencing methodologies.
Our cohort of 107 DCM patients had similar baseline characteristics to the cohort of the European INHERITANCE project (which did not include Portugal), published in 2015 by Haas et al.,10 although we had fewer symptomatic patients (49% of our patients were in NYHA class I vs. 28% of the European cohort) and a lower proportion of heart transplant recipients (10% vs. 20%).
We found rare variants in approximately one fourth of patients. This proportion is similar to that of a study by Lakdawala et al.,26 lower than the results of a recently published Finnish cohort16 (35% of patients presented with pathogenic or likely pathogenic variants), and significantly lower than the proportion of positive molecular results obtained by Haas et al.,10 in which variants causing or associated with DCM were described up to 73% of patients. This lower diagnostic yield might be related to the fact that other genes potentially related to the disease were not included,26 most importantly the titin gene (TTN), which is presumed to account for up to 14-25% of all DCM cases27 and for the majority of variants found in the INHERITANCE project10 and in the Finnish cohort,16 but may also be related to a different degree of stringency for the classification criteria of variants. These differences are even more pronounced taking into account that we considered both variants that are likely pathogenic and those of uncertain significance. We decided to do so because most of the variants identified, although predicted to be functionally significant by in silico prediction tools, were classified as of uncertain significance mainly because they were novel and originally reported in this study, and familial segregation analysis was not informative.
Although without statistical significance, we found a higher proportion of patients with rare variants in familial DCM patients, which is in line with other studies.10,26 Even so, a genetic origin is presumed in the idiopathic cases, because of the occurrence of the novo variants, incomplete penetrance, limited number of genes analyzed and small families.
The majority of variants were found in sarcomeric genes, as in other reports,10,26 most of them in MYBPC3 (10/31). MYBPC3 variants are frequent in patients with HCM, representing 30-45% of all HCM-associated variants.28 In patients with DCM, MYBPC3 variants are thought to account for approximately 2-4% of cases,6,27 a lower percentage than in our cohort. As most of these variants (8/10) had previously been described in association with HCM, one might hypothesize that the DCM phenotype represented end-stage HCM (‘burnout’ physiology). We consider this hypothesis less likely in view of the follow-up of these patients previous to the inclusion in the study, the relatively young age of presentation, and family history of DCM in all but one patient.
In our study we could not define clear genotype-phenotype correlations for patients with a rare genetic variant, or in patients with specific variants. Although TNNT2 variant carriers appear to present at a younger age, in agreement with a recent meta-analysis by Kayvanpour et al.,27 the presence of a higher left ventricular ejection fraction in these patients and in MYH7 variant carriers was not confirmed. Also, we found no statistically significant differences in gender predominance, conduction disease, ventricular or supraventricular arrhythmias or the occurrence of heart transplantation, probably due to the small number of patients with gene-specific variants. Even so, two of three patients with PLN and LMNA variants had previously received ICDs and all patients with LMNA variants underwent heart transplantation (two after enrollment in the study), in line with the observation of a higher prevalence of heart transplantation and ventricular arrhythmias in carriers of LMNA and PLN variants.10,27
The presence of more than one variant in DCM patients is in agreement with the study by Haas et al.,10 in which more than 38% of patients had two variants and 13% had three or more. However, it is not known whether the presence of two or more variants in a single individual alters the phenotypic expression. Some authors argue that some low-penetrance variants might promote a disease phenotype only in the presence of other phenomic, genomic or other epigenetic factors.6 This could explain the occurrence of DCM in the family illustrated in Figure 2A, in whom the phenotype is only expressed at a young age in the individual with two variants of uncertain significance, and the presence of just one variant is not associated with the expression of DCM. Additionally, the presence of multiple variants could be related to a more severe phenotype and clinical course, as has been described for double heterozygotes for LMNA and TTN variant carriers with respect to age of end-stage HF and heart transplantation.29 In fact, in our study, two patients with multiple variants presented a particularly unfavorable clinical scenario, with end-stage HF and subsequent heart transplantation.
Determining whether a rare genetic variant is responsible for the disease is the most critical aspect of genetic testing. Interpreting whether a genetic variant is pathogenic is increasingly complex due to the large number of rare variants in the human genome and the advent of NGS, which enables analysis of extended gene panels. In many studies, relative loose frequency criteria have been used to ascertain variant pathogenicity.10,30 In particular, as reported by Akinrinade et al.,16 the European INHERITANCE project10 reported that 73% of the patients had ‘known’, ‘likely’, or ‘potential’ disease-causing variants (46% of the patients with ‘known’ disease-causing variants in cardiomyopathy or channelopathy genes, but only 16% of those with ‘known’ DCM variants), and variant interpretation was based mainly on variants reported in HGMD, which is known to contain variants classified as pathogenic, although some are increasingly being identified as benign polymorphisms. Similarly to our study, Akinrinade et al.16 used the ExAC database,29 which includes exomes of >60 000 individuals (10 times more than the Exome Sequencing Project database used by Haas et al.10) as the reference population database. Additionally, these authors randomly selected 10 variants from Haas et al.’s study classified as potential disease-causing (classes Ia-III) variants and ascertained their frequency in the ExAC database, finding that 80% of the variants were present in over nine (allele frequency >0.015%) subjects in the ExAC database with an average of 263 carriers (allele frequency >0.44%) in the database, making these variants highly unlikely to be disease-causing. In our study, we applied the criteria in the ACMG/AMP guidelines,22 which may be rather more stringent than the criteria applied to date, possibly leading to a larger proportion of variants being categorized as of uncertain significance, in order to reduce the number of variants being reported as ‘causative’ of disease without sufficient supporting evidence for that classification.
Recently, Walsh et al.32 compared sequence data of 7855 individuals with a clinical diagnosis of cardiomyopathy, including 559 with DCM, with 60 706 reference samples from the ExAC database. In clinical cohorts, variants in TTN were the most common, with the prevalence of rare variants in MYH7, LMNA, TNNT2 and TPM1 being modest, but significantly enriched compared with ExAC. However, there was modest or no significant excess variation in other commonly DCM-associated genes, like MYBPC3, with the authors arguing against a causative role for this gene in DCM pathogenesis, which challenges some widely held conceptions10,33 and our own results. Moreover, in genes supported by an excess of pathogenic and likely pathogenic variants, even variants of uncertain significance were seen to be enriched when compared to ExAC, suggesting that clinical laboratories may be overly conservative.32
All these findings underscore the need to continue exploration of genetic mechanisms and determinants of DCM and to establish standardized and reliable criteria for defining the pathogenicity of genetic variants. This will enable accurate interpretation of genetic testing, although it should be kept in mind that as new knowledge becomes available, the classification of previously identified variants may change.
LimitationsThe major limitations of our work are the relatively small number of patients included and the limited panel of analyzed genes, particularly the non-inclusion of TTN, which is currently known to account for a large percentage of all DCM cases. Additionally, structural variants were not considered in this study, as structural variants were described in association with DCM34–37 after the study design was finalized and for some of the genes with the strongest association (such as LMNA) a multiplex ligation-dependent probe amplification kit was not available. Although structural variants are associated with a very small percentage of DCM cases (about less than 1%, according to the current literature), it would be valuable to search for this type of variant in a future project.
Another limitation is related to family segregation analysis, which was performed in very few cases, due to the small size of the families, unavailability of some family members for clinical and genetic assessment, and lack of data.
ConclusionsThis is the first study to address the genetic background of Portuguese patients with DCM and reflects the complexity and diversity of DCM genetics.
For better interpretation of the pathogenicity of the variants found and their causative role in DCM, careful phenotypic assessment of patients and their families and molecular cascade screening are crucial. Only with further advances in diagnostic techniques and improved understanding of biological mechanisms can we gain further insight into genotype-phenotype correlations and be able to use genetics for risk stratification, as well as to develop appropriate strategies for DCM prevention and management, ultimately improving the care of patients and families with DCM.
Funding sourcesThis work was supported by the Portuguese Government's Foundation for Science and Technology [PTDC/BIM-MEC/0650/2012].
Conflicts of interestThe authors have no conflicts of interest to declare.
We thank the FATIMA (Portuguese study of familial dilated cardiomyopathy) investigators. Dr. Adriana Belo, from the Portuguese Society of Cardiology, provided statistical support.